
Yves Gaetan
@yvesgnt
PhD Student at @IBMResearch and @TUeindhoven (@molecularML). Working on language models for protein design and optimization 🇮🇹 🇨🇲🏳️🌈
ID: 1343315401854373899
27-12-2020 21:58:54
148 Tweet
306 Takipçi
357 Takip Edilen

Are you passionate about the untapped potential of #AI in drug discovery? Then, check out our openings Molecular Machine Learning: 1 PhD & 1 Postdoc positions, funded by European Research Council (ERC)! You will be hosted at the vibrant TU Eindhoven & @ICMStue. See below! 👇 (1/4)


Are you a master student? Eager to learn about #QuantumComputing? Combining optimization (#OptimalTransport) with #MachineLearning & applying to cell biology sounds interesting? Come join us IBM Research for an interdisciplinary thesis! Apply: ibm.biz/msc_qdd Please RT!

Happy two-year anniversary to the Molecular Machine Learning team! 🎂 What a journey it has been so far — filled with immense satisfaction, cool collaborations, and incredibly talented people. A toast to stretching the limits of #AI in drug discovery and chemical biology 🥂


🚀 Exciting news! Our paper on GT4SD has been published in npj Journals Computational Materials! Learn more about it here: doi.org/10.1038/s41524…. Try it out: GitHub - github.com/GT4SD/gt4sd-co… Demo apps on HF Spaces - huggingface.co/GT4SD

Explore our latest paper on designing catalysts using deep generative models and computational data. Discover how an RNN-VAE can generate new catalyst candidates for the Suzuki coupling. doi.org/10.1039/D2DD00… IBM Research LIAC at EPFL Alain Vaucher Philippe Schwaller (he/him) Teodoro Laino

Very excited our Preprint “Leveraging Infrared Spectroscopy for Automated Structure Elucidation” is out. First of its kind model: Automatic Structure determination from IR spectra! Top-1 Accuracy of 45%! Alain Vaucher Teodoro Laino IBM Research Paper: ibm.biz/ir_to_struc 🧵⬇️


Our review on structure-based #drugdiscovery is out! We cover it's different facets, ranging from drug-target interaction prediction and binding site prediction, all the way to de novo drug design. Finally, we look at the challenges ahead 🔮 Molecular Machine Learning #ML #AI #chemistry

📢 If you are attending ACS Fall 2023 in San Francisco and you want to know more about #LanguageModels applied to material design, stop by Nob Hill A - Marriot Marquis at 10:25AM today IBM Research


If you are #ACSFall2023 and want to learn how we can utilize AI to predict the right solvent for a chemical reaction, check out my talk today at 3:10Pm at Nikko II - Hotel Nikko or read the preprint doi.org/10.26434/chemr… IBM Research LIAC at EPFL

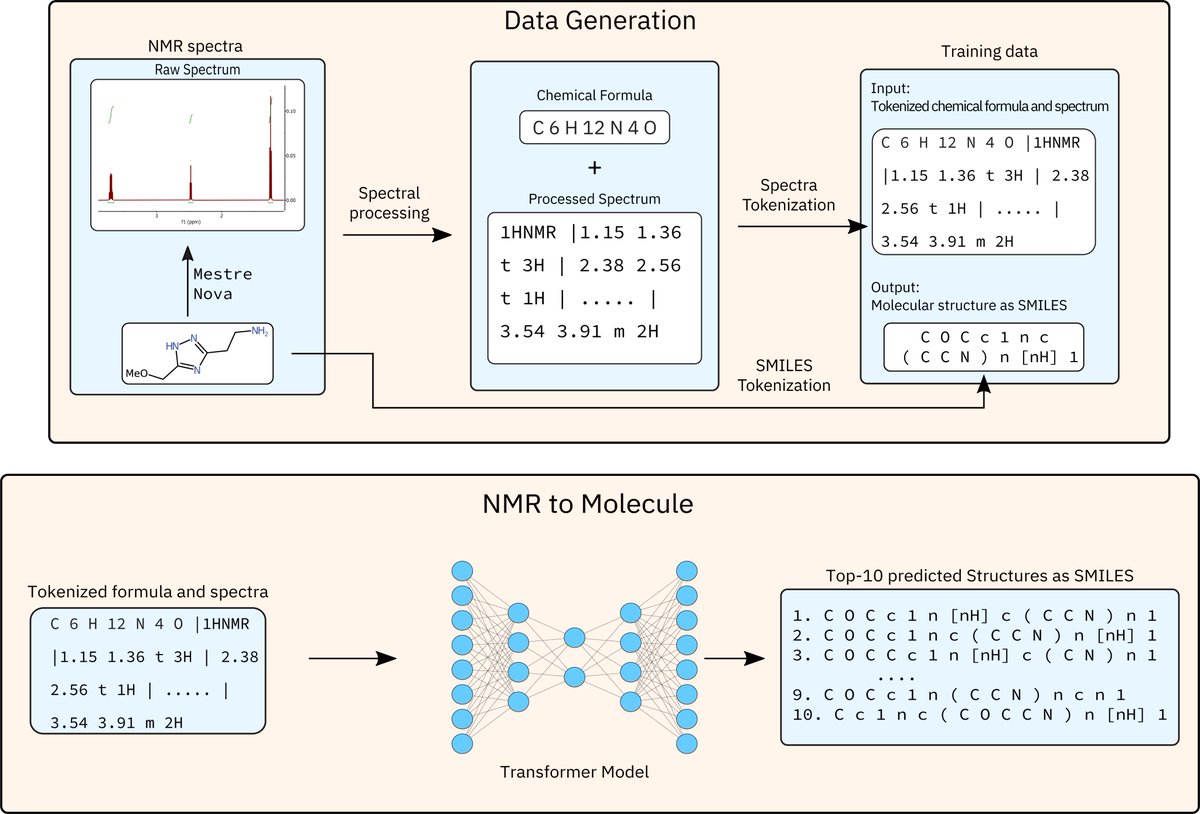
Check out our Preprint in ChemRxiv on "Learning the Language of NMR: Structure Elucidation from NMR spectra using Transformer Models". We train models to directly predict the molecular structure from NMRs! Paper: ibm.biz/nmr_to_struc Alain Vaucher Federico Zipoli IBM Research

Ana Ortiz and I just released an exciting preprint on “Machine learning-guided high throughput nanoparticle design” with Roy van der Meel Francesca Grisoni Lorenzo Albertazzi doi.org/10.26434/chemr… Molecular Machine Learning Nanoscopy for Nanomedicine lab #ML #nanomedicine #nanoparticles #drugdiscovery #AI A small 🧵 below👇

📢 Open postdoc position on protein representation learning for drug discovery! 📢 Funded by European Research Council (ERC) 🇪🇺 Join our vibrant team Molecular Machine Learning+TU Eindhoven to push the boundaries of #AI for #drugdiscovery. 💪🏻 Apply before October 31st!👇 jobs.tue.nl/en/vacancy/pos… Please RT 🙂


Super thrilled about the work of Derek van Tilborg on active #deeplearning for drug discovery 💪🏻 We address a broad range of questions on how to use active learning to explore the chemical space.🚀 It turns out active deep learning is quite powerful, even with little data!👇






