Ye Zheng
@yezhengstat
Assistant Professor@MD Anderson Bioinfo and Comp Bio (joint Systems Biology); NIH/NHGRI K99; Statistics; Epigenomics and 3D Genomics; Cancer Studies
ID: 720078151011995648
http://www.stat.wisc.edu/~yezheng 13-04-2016 02:36:24
763 Tweet
1,1K Followers
1,1K Following




🧬 Researchers at Fred Hutch Cancer Center have uncovered an overlooked mechanism driving aggressive breast and brain tumors. Learn more about the 64 ancestral genes investigated by the Henikoff lab: bit.ly/3XIke4Y Science Magazine abstract: bit.ly/4iGJAbtResearc…


Excited to share another collaborative study with Dr. Joe Costello - Intratumor heterogeneity of 3D genome organization in primary glioblastoma samples. Led by Qixuan and Juan, and congrats to the whole team! Lurie Cancer Center Northwestern Feinberg School of Medicine UCSF Neurosurgery science.org/doi/10.1126/sc…




Our spatial FFPE-ATAC-seq is now published in Nature Communications! With extensive optimizations, it performs well across diverse tissues, unlocking the vast FFPE archive for spatial epigenomics. Big congrats to Pengfei Guo Pengfei_Guo, Yufan Chen, and the team! nature.com/articles/s4146…

📢 Now live at nature Our team discovered that PPP2R1A mutations are linked to improved response to immunotherapy in ovarian clear cell carcinoma. Yibo Amir Jazaeri Rugang Zhang Lab MD Anderson Cancer Center nature.com/articles/s4158…

Our scMicro-C paper is out in Nature Genetics! Huge thanks to the reviewers for their valuable insights, to SunneyXie for unwavering support, and to fantastic collaborators Jiankun Zhang & Longzhi Tan. Thrilled to share this work!




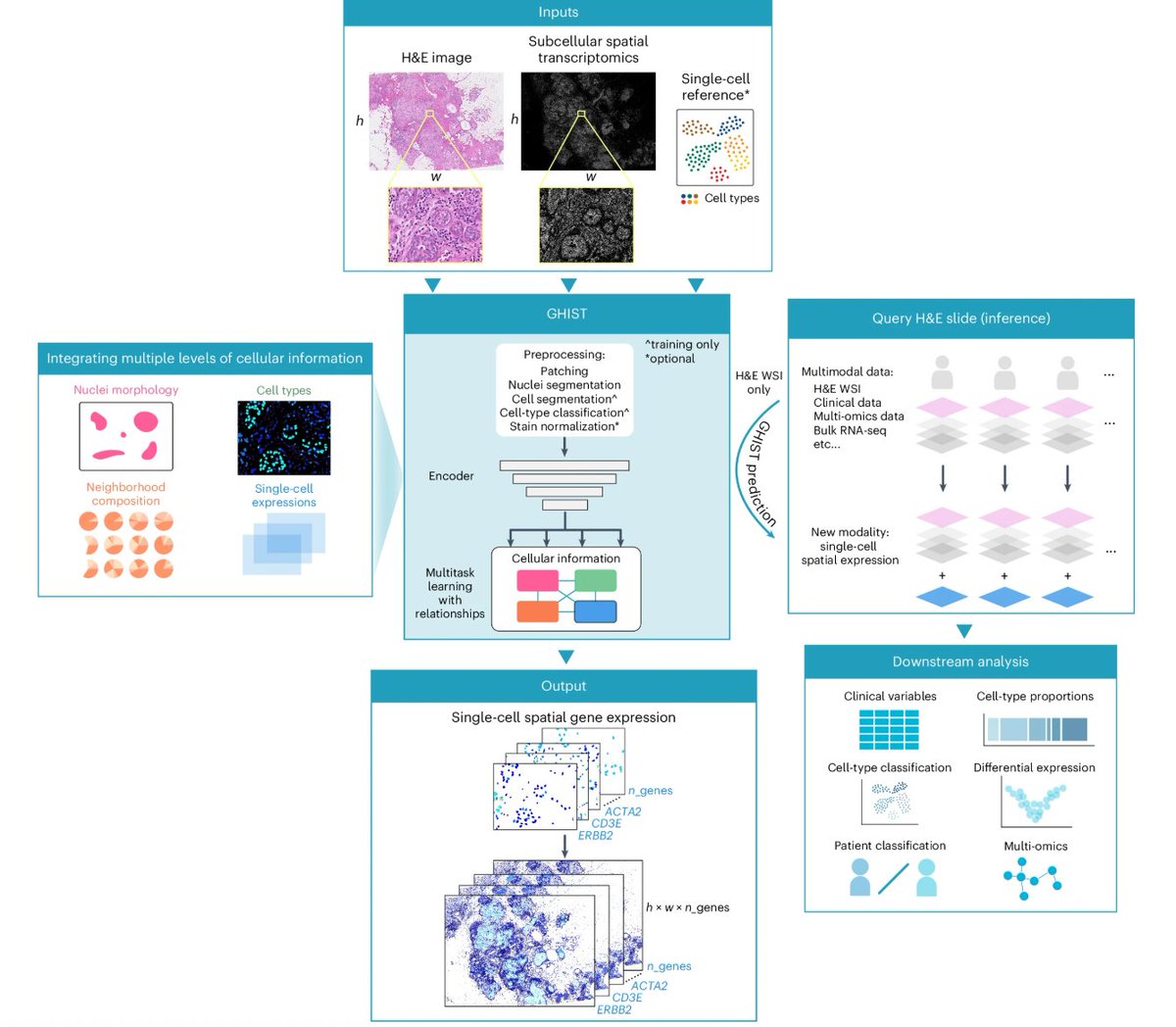
It was a pleasure to write this piece in Nature Methods on GHIST & iSCALE. These two methods transform H&E histology into molecular maps, predicting gene expression at single-cell and whole-slide resolution, expanding spatial omics at scale!

[1/10] We’re excited to introduce CytoTRACE 2 — a major upgrade for inferring developmental potential from single-cell RNA-seq across datasets. Nature Methods Led by Minji Kang in Aaron Newman Lab. 📄 Paper: nature.com/articles/s4159… 🔗 Shareable link: rdcu.be/eMTVO

Thrilled see our paper out in nature ! 🤩 Wonderful collaboration with GonçaloCasteloBranco. Kudos to my amazing postdoc Di at Yale Engineering and Leslie Kirby from Goncalo's lab Karolinska Institutet. Huge team science with Yale School of Medicine UC San Francisco A*STAR nature.com/articles/s4158…