
William DeWitt
@wsdewitt
Assistant Professor @UW @uwgenome
Previous: JSMF Fellow @Berkeley_EECS
♡: Computational biology, evolutionary dynamics, quantitative immunology
ID: 632482420
https://wsdewitt.github.io 11-07-2012 01:42:51
966 Tweet
776 Followers
1,1K Following


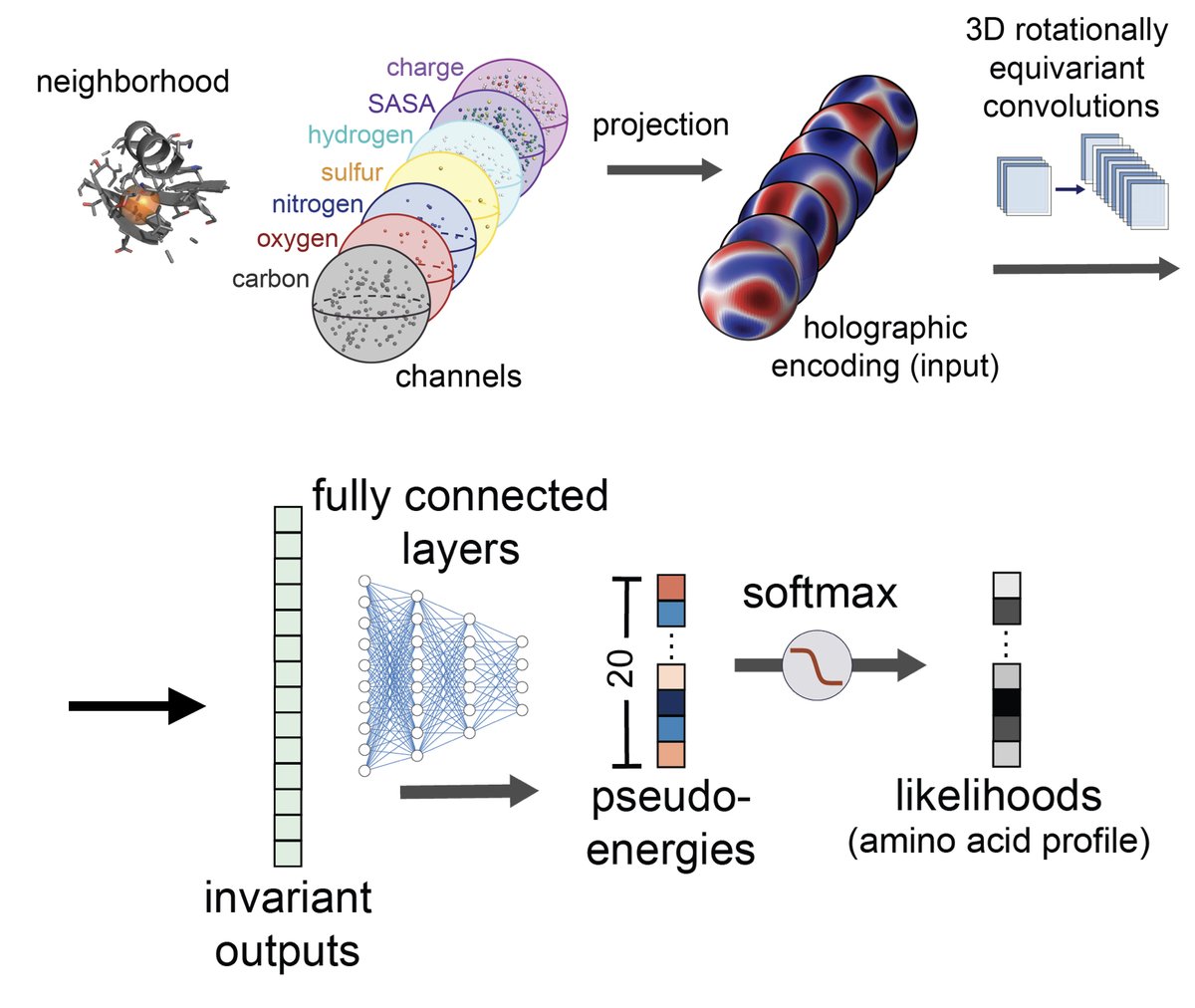
Check out our new preprint (led by Mike Pun University of Washington) on 3D rotationally equivariant model of protein structure micro-environments, with a holographic CNN (H-CNN): doi.org/10.1101/2022.1…

Looking forward to Sebastian Prillo’s talk tomorrow morning 🍿


I had some snow-related airport delays in Seattle last week, but am very thankful for the chance to sneak in a cheeky powder day. So fun! 📷 by David Bacsik



Andrew Kern and I are organizing a symposium at #SMBE23 on machine learning and evolutionary genomics. We are delighted to have Yun S. Song and Sohini Ramachandran as our invited speakers. Should be a blast! Abstract submissions are due on March 15. We hope to see you in Ferrara!

Recently Richard Wang Matthew Hahn et al inferred past human generation times from changes in the mutation spectrum over time. We think this approach is heavily biased by uncertainties in allele age and de novo mutation rate estimates. See biorxiv.org/content/10.110… and below:



Last year, a team led by @tomsasani and I found that germline mutation rates and spectra differ between C57BL/6 and DBA/2J lab mice. We mapped the C>A mutator phenotype to the DNA repair gene Mutyh. Now with Aaron Quinlan we show there's more to the story! biorxiv.org/content/10.110…




Very glad to see our CherryML method published online in Nature Methods. The published version has updated and expanded results, so please check it out. doi.org/10.1038/s41592… (1/3)

Today out in Nature Reviews Genetics: our review on how evolutionary control should be devised to improve synthetic biology, molecular design, and drug and vaccine design against pathogens. with Michael Lässig, & Ville Mustonen nature.com/articles/s4157…



