
Oleg Kovalevskiy
@ovkovalevskiy
Structural Biologist
ID: 1570776316202438657
16-09-2022 14:07:49
26 Tweet
103 Takipçi
86 Takip Edilen


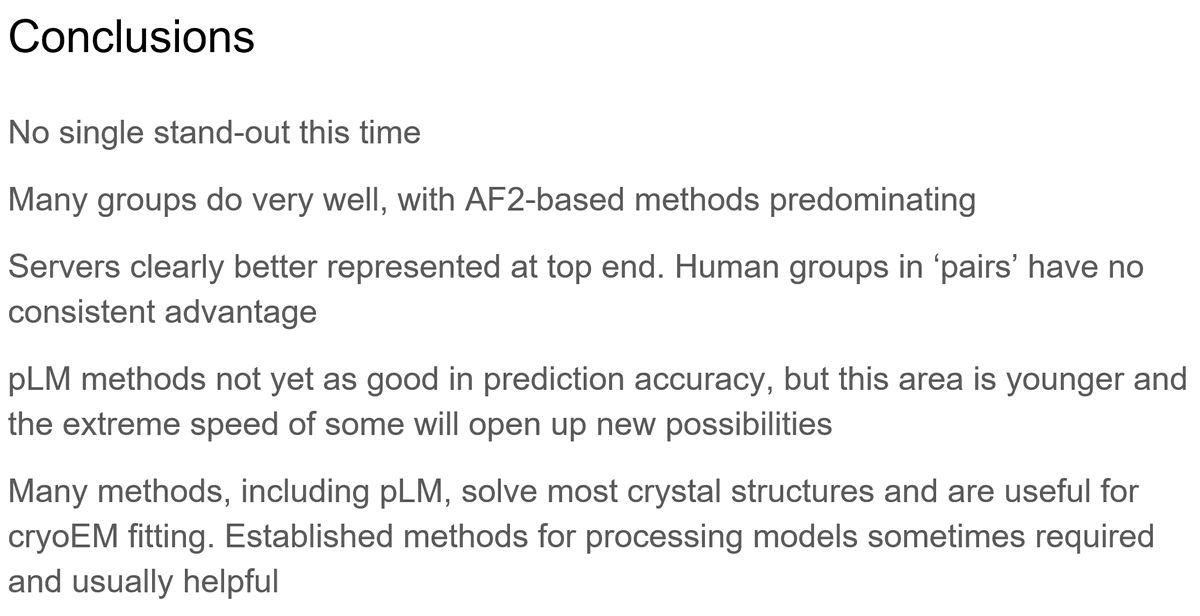
A pleasure and a privilege to be assessor again at #CASP15. A mix of spectacular success and the occasional intriguing failure. Looking forward to hearing from the predictors and discussing future directions. Thanks @AJSimpkin Ronan Keegan Shah Mes David L Murphy Filo and Luc.





So happy to see our work going live together with that of Pedro Beltrao and @thesteinegger today @nature! Biozentrum, University of Basel Universität Basel SIB SWISS-MODEL link: nature.com/articles/s4158… link: nature.com/articles/s4158…


Today in partnership with Isomorphic Labs, we're sharing progress on the next generation of #AlphaFold. 🧬 Our new model greatly expands coverage of structure prediction beyond proteins to other key biomolecular classes. How could it help scientists? 🧵 dpmd.ai/alphafold-prog…








The #AlphaFold 3 model code and weights are now available for academic use. We Google DeepMind are excited to see how the research community continues to use AlphaFold to address open questions in biology and new lines of research. github.com/google-deepmin…








