
Melanie Kirsche
@melanie_kirsche
Bioinformatics Scientist at Roche | PhD from @JohnsHopkins | she/her 🏳️⚧️
ID: 1170080572342800384
06-09-2019 21:05:30
33 Tweet
184 Takipçi
74 Takip Edilen

Very excited to announce: "Jasmine: Population-scale structural variant comparison and analysis" by Melanie Kirsche Rachel Sherman, PhD Bohan Ni Sergey Aganezov and Gautam Prabhu. biorxiv.org/content/10.110…

I'm honored to be a part of this exciting work exploring some of the improvements from using CHM13 as a reference compared to GRCh38! Thanks to the other co-first authors @sz_genomics Stephanie Yan Daniela C. Soto Sergey Aganezov, as well as everyone else who contributed!

Thank you to Michael Schatz and to the rest of my committee! I'm very excited to finally be a doctor!



Scientists from all over the world worked to complete the project, including many early-career researchers and trainees! Some of these incredible researchers include: HHMI UC Berkeley @epitopic @roneilllab @uconnresearch UCDavis News Service UC Santa Cruz UC Santa Cruz Genomics Institute Uniklinik Münster


So proud and excited for our #T2T work on "A complete reference genome improves analysis of human genetic variation"! Im enormously grateful to the whole team for their tireless work and support Bloomberg Distinguished Professors JHU Computer Science Cell, Molecular, Developmental Biology, Biophysics science.org/doi/10.1126/sc…

Latest paper is out in Cell Genomics from the Genome in a Bottle crew! "Benchmarking challenging small variants with linked and long reads" cell.com/cell-genomics/… Andrew Carroll Fritz Sedlazeck @sz_genomics Bohan Ni Melanie Kirsche Sergey Aganezov Giuseppe Narzisi et al

Incredibly proud to have been selected with my good friends Adam Phillippy Karen Miga and Evan Eichler on behalf of the entire T2T consortium!!! TIME #time100 Johns Hopkins University Bloomberg Distinguished Professors time.com/collection/100…

Check out Ariel Gershman 's flash talk at #AGBT22 at 11:30 on her #T2T epigenetic work! Her poster is #419 "Long reads reveal epigenetic patterns from telomere to telomere"

So excited to be giving my first in-person conference talk at #AGBT22 in just over an hour! Come to the comp bio session to hear all about my work on building a long-read SV analysis pipeline with Michael Schatz Sergey Aganezov Bohan Ni and many others!

Come check out Melanie Kirsche 's talk at #AGBT22 Computational Biology session at 7:50 "Jasmine: Population-scale structural variant comparison and analysis"!

Very excited to be presenting at #AGBT22 on how #T2T-CHM13 improves human genetics analysis across the board! Stop by my talk at 11:20am tomorrow to hear more about this collab with Sergey Aganezov, Daniela C. Soto, Melanie Kirsche, @sz_genomics, and many others on the T2T team 🧬👀



I’m very excited to be joining the bioinformatics team at Variant Bio, a company which studies the genetics of people and groups with exceptional health-related traits to inform drug development. More info here: variantbio.com

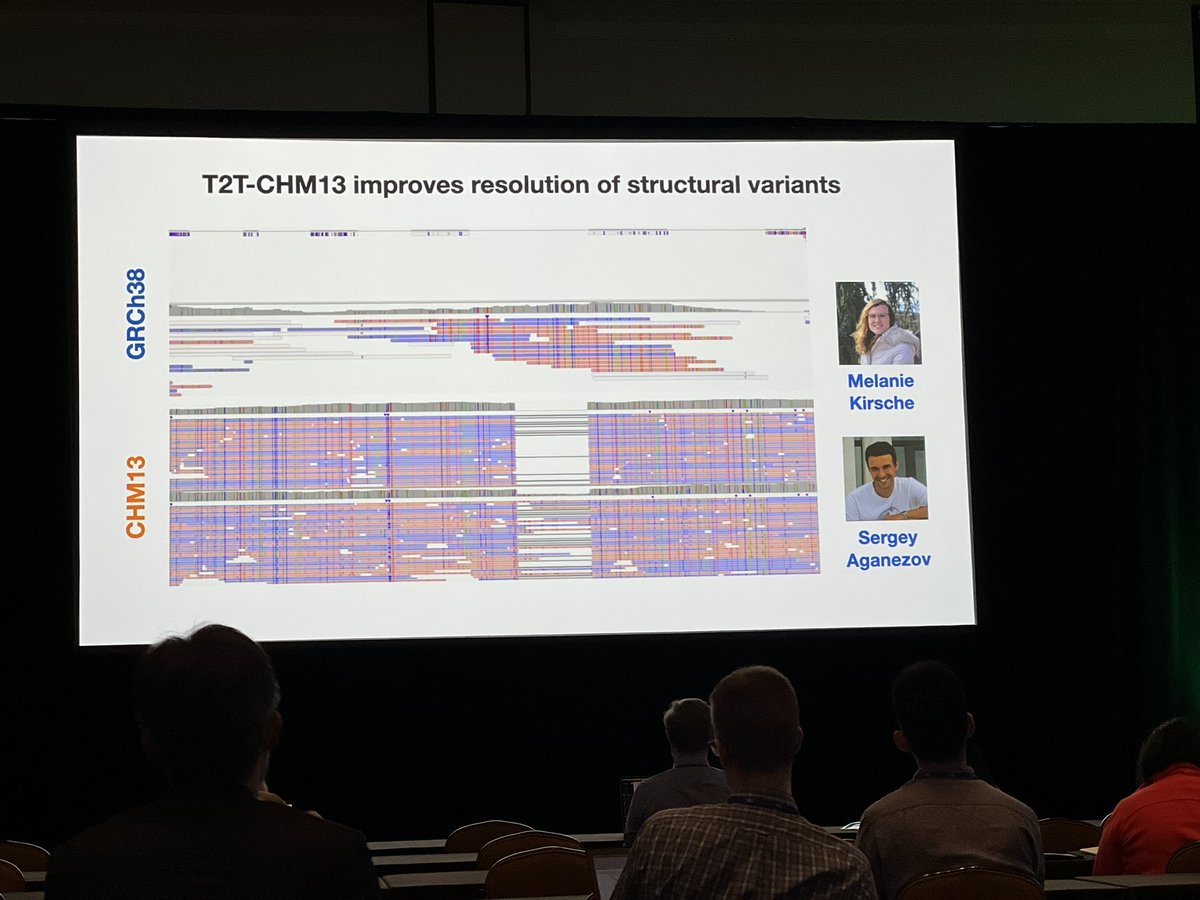
Using the T2T reference genome (CHM13)vs GRCv38 improves alignment and detection of structural variants for Oxford Nanopore and PacBio data from NY Genome Center data #AGBT22


Super excited to announce "Jasmine and Iris: population-scale structural variant comparison and analysis" by Melanie Kirsche, Guautam Prabhu, Rachel Sherman, PhD, Bohan Ni, Alexis Battle & Sergey Aganezov. Bloomberg Distinguished Professors JHU Computer Science nature.com/articles/s4159…

Very proud to have contributed to this study with Mark Gerstein rozowsky Sergey Aganezov Melanie Kirsche Fritz Sedlazeck Rachel Sherman, PhD Ruibang Laurent Luo Srividya Ramakrishna et al. Shows the importance of gene regulation analysis by mapping functional data to personalized genomes of the individuals


