Marek Mutwil 🇺🇦
@labmutwil
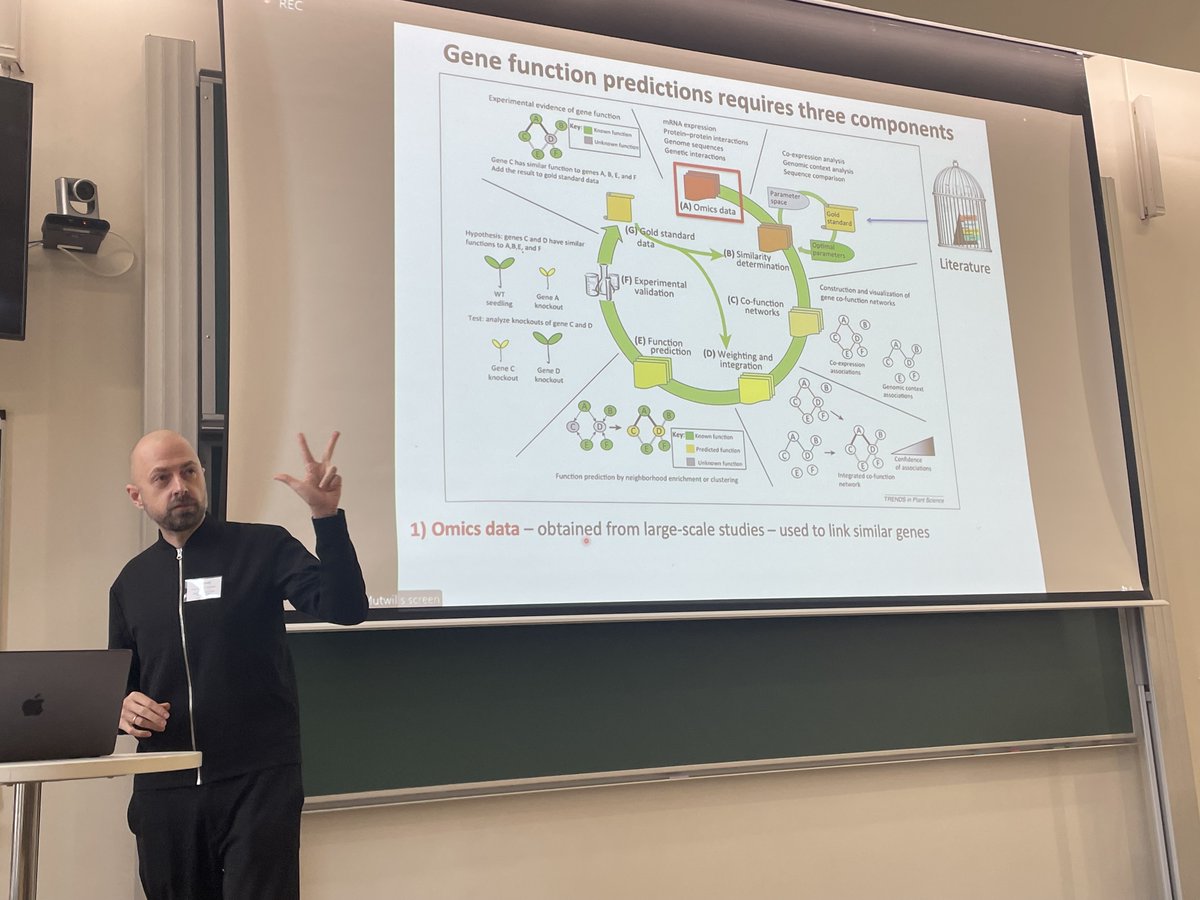
Aim: improve gene function predictions of the plant kingdom to produce more climate change-resilient and nutritious crops.
ID: 2865395000
https://mutwillab.com/ 19-10-2014 16:49:53
1,1K Tweet
1,1K Takipçi
700 Takip Edilen



#PlantWorkhop at @spsplantsciences.bsky.social Marek Mutwil (Marek Mutwil 🇺🇦) "Gene function predictions requires three components.. More omics data, More Gold Standard data and a better function prediction method"