GNPS - UC San Diego
@gnps_ucsd
Online mass spec analysis platform that enables global visualization and interpretation based on crowd sourcing. #molecularnetworking #massspec
ID: 2521441274
http://gnps.ucsd.edu 24-05-2014 22:04:41
2,2K Tweet
2,2K Takipçi
68 Takip Edilen


Ming Wang GNPS - UC San Diego mzmine OpenMS Team Anyone interested in using MassQL in UniDec can find it in the experimental tab of the main menu. It’s a great way to search for specific features in your complex peak list!



Oscar Yanes Justin van der Hooft depends on how you look at it. In sheer data probably yes. But the groundwork (like MS technologies, GNPS - UC San Diego etc) usually takes most work and energy. Think of the sequencing technologies.

New paper from Yasin El Abiead! He even used MassQL to search for multimeric metabolites across all public mass spec data.


We are very excited to welcome Ming Wang, a new UCR Computer Science and Engineering faculty. His lab is actively recruiting undergraduates and graduate students so if you're interested in bioinformatics and understanding the chemical space around us, check out the opportunities: cs.ucr.edu/~mingxunw/oppo…

Lars highlights how SynCom influences SM levels produced by Pseudomonas protegens = cool chemical data (LC-MS) combined with GNPS - UC San Diego analysis and beautiful MS imaging of duo colonies demonstrate multispecies interaction (MSI funded by Novo Nordisk Foundation) #LipopeptideLiege


Out now! 🥳 We synthesized silicone (PDMS) foams to sample #Air gas&particles + performed #NTA by GC & LC-HRMS Orbitrap to get the full picture! Proud and excited to share our work Environmental Science & Technology Journals Letters Jon Martin Bénilde Bonnefille Hongyu Xie GNPS - UC San Diego life(Lite) for #Exposomics

Join Anelize Bauermeister @amcaraballor Tiago Leão to learn how to GC/MS Molecular Networking in Portuguese! youtube.com/watch?v=WLk9kL… youtube.com/watch?v=gVJPwt…


Our team is recruiting a post-doc in analytical chemistry for 2 years on the moss Physcomitrium patens at IJPB in frame of a collaborative project about #strigolactones IJPB Isabella and US2B Nantes Université. Contact [email protected] Please RT


The preprint is out! "UmetaFlow: An untargeted metabolomics workflow for HT data processing& analysis". A huge thank you to the OpenMS Team, the NPGMgroup, and especially to A. Walter, O. Alka, J. Pfeuffer, Timo Sachsenberg & Oliver Kohlbacher - Now @okohlbacher.bsky.social! 🙏🥳 So grateful. doi.org/10.26434/chemr…

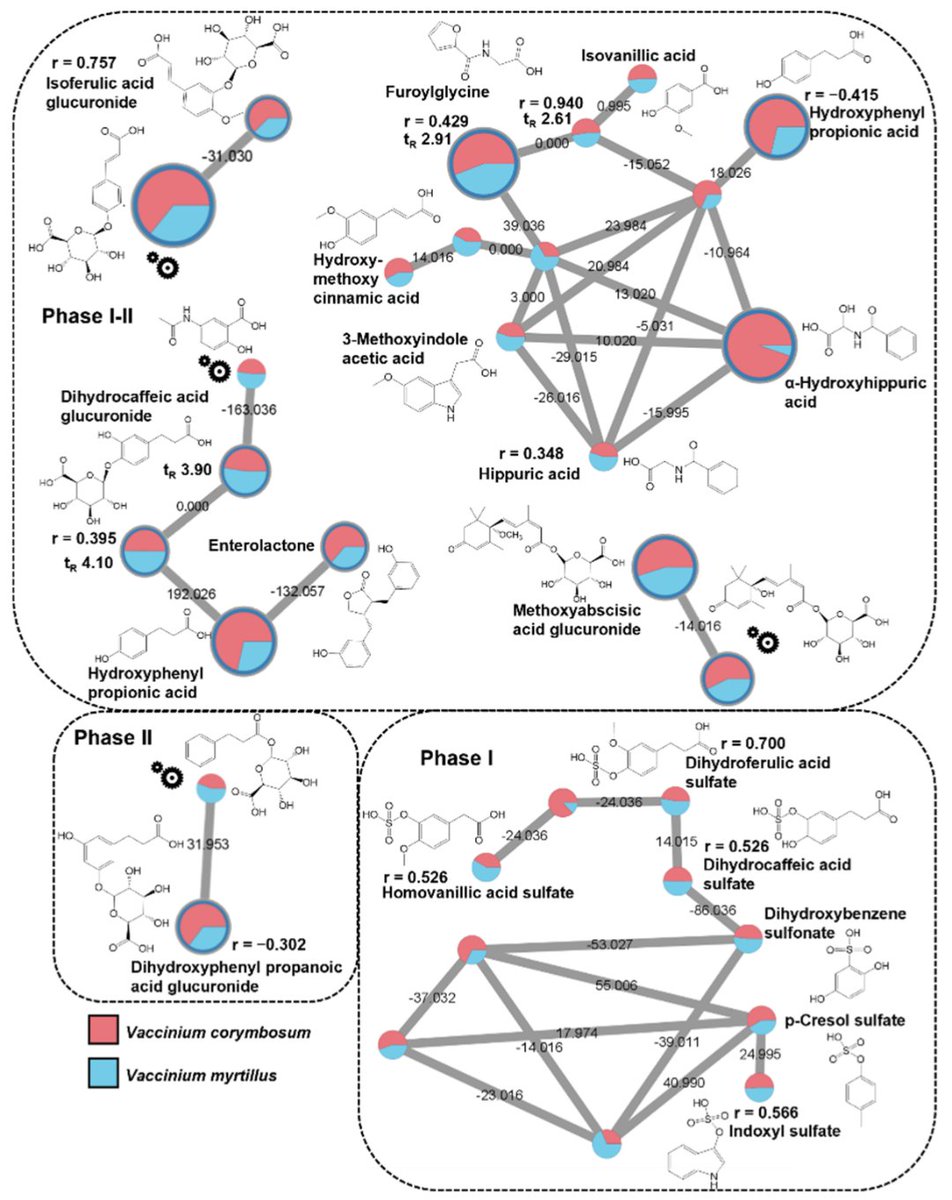
The combination with #feature based #molecular #networking GNPS - UC San Diego #FBMN allowed us to probe #berry derived #urine metabolites in the #metabolomics #profiles! 😎 Further, we could speculate on their Phase I or II origin due to available #time series and #quantitative info!

Great to see more metabolomics data being shared thx to many impressive efforts @metaboLights @metabolomicsWB @gnps_ucsd Hope this article can be a starting pt of discussion for Nature Cell Biology readers to facilitate data re-use. Nice work Ethan Stancliffe! nature.com/articles/s4155…

Very happy to share with you our collaborative work with Publications MNHN 🐝 Marine Drugs MDPI that melted together #computationalmetabolomicstools such as #MASST GNPS - UC San Diego shorturl.at/tzW78



Next stop on our Workshop Europe tour: Zurich hosted by Nicola Zamboni. Corinna and I look forward to discussions and meeting our colleagues from the Swiss Metabolomics Society at ETH Zurich. We will present work by mzmine, GNPS - UC San Diego, SteffenHeu, Ming Wang, and others.