Claudia Driscoll
@driscoll_cl
Postdoc in the Hie Lab at @arcinstitute and @Stanford | Prev @BiochemOxford
ID: 1346749840806600704
06-01-2021 09:26:29
3 Tweet
38 Takipçi
314 Takip Edilen

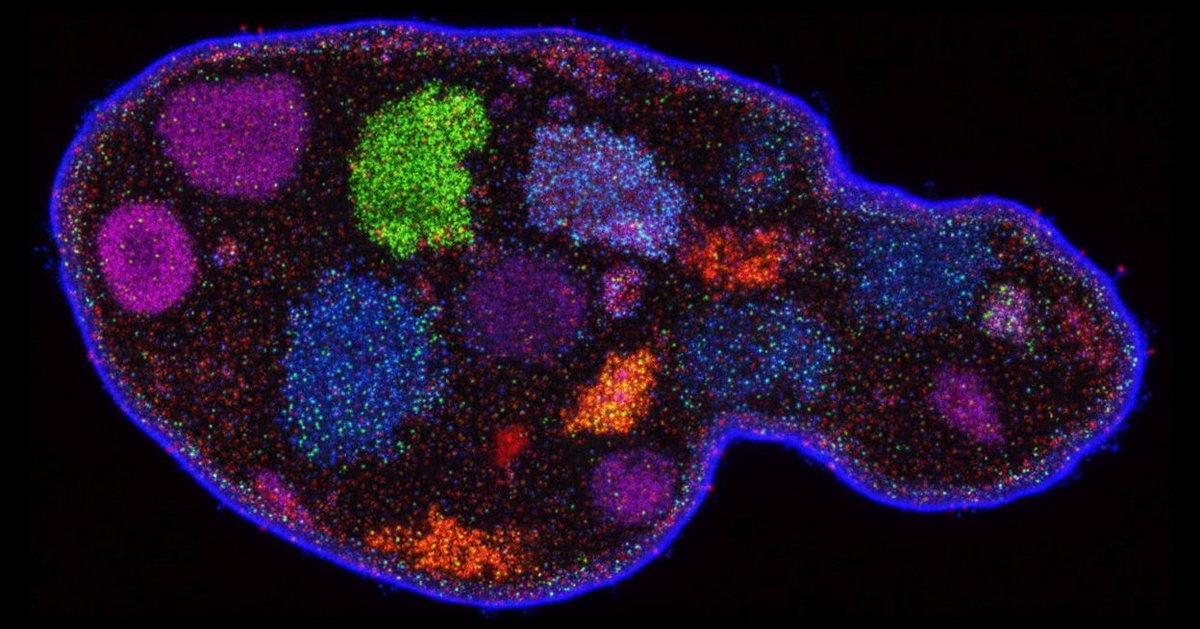
New from Arc Institute: the first functional AI-generated genomes. This is a conceptual breakthrough, but it may also unlock new strategies for combating antibiotic resistance.

Evo-designed genomes are here!! HUGE congrats to Samuel King for fearlessly bringing this project to life. Check out the thread to learn more! ⬇️




In another preprint from the @brianhie Lab and Xiaojing Gao, they introduce Germinal, a generative AI system for de novo antibody design. Germinal produces functional nanobodies in just dozens of tests, making custom antibody design more accessible than ever before.


Today, we report Germinal, a method for efficient de novo antibody design, with Santiago Mille and Xiaojing Gao. Germinal achieves success rates of 4-22% across diverse epitopes. We make the work fully open, without doing lame things like posting a preprint without methods. 🧵



Super excited to share what we’ve been working on in collaboration with Xiaojing Gao over the past few months on de novo antibody design. Check out this great thread by our team lead Santiago Mille highlighting the technical aspects of the pipeline!






Excited to share that my PhD thesis work is out in Science Magazine today. We demonstrate robust rearrangement of the human genome using bridge recombinases, performing programmable insertions, excisions, and inversions at megabase-scale.


Combine multimer structure prediction and an antibody language model to design de novo antibodies with nanomolar binding affinity. Santiago Mille John Wang Talal Widatalla Claudia Driscoll Haoyu Dai Xiaowei Zhang Xiaojing Gao Brian Hie