
robin
@_robin_meyers_
PhD student. formerly DepMap@Broad. genomics, systems and cancer bio, dna damage and genomic instability, genetic interactions, chromatin/gene regulation
ID: 1902874531
https://scholar.google.com/citations?user=XrzHDx4AAAAJ 25-09-2013 04:38:39
29 Tweet
105 Takipçi
547 Takip Edilen


Identification of WRN as a novel synthetic lethal target in MSI cancers! Congratulations to our team Ed Chan, Tsukasa Shibue, Francisca Vazquez, Adam Bass The Cancer Dependency Map @BassLab_DFCI disq.us/t/39rxsuo

Proud to announce that the capstone project of my thesis work in Kadoch Lab, is avail. online today in @nature_genetics: nature.com/articles/s4158… 1/n




Advice from Daniel MacArthur to his team following yesterday's Ioannidis lecture on reproducibility



Genome-wide #CRISPR screens of cancer cells affected by bias across CNAs. Our paper is out! Aviad Tsherniak Broad Institute ncbi.nlm.nih.gov/pubmed/27260156

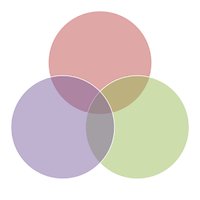
The largest dataset of cancer genetic dependencies: More than 970 RNAi screens in 713 cell lines from Broad Institute Achilles, Novartis DRIVE and Marcotte et al., 2016 processed together by the new DEMETER2 method. biorxiv.org/content/early/… The Cancer Dependency Map bioRxiv


In this paper, robin and I take a look at how patterns of co-dependency in cancer cell lines teach us about protein complex biology. Credit to a fantastic team Aviad Tsherniak, Ann Sizemore Blevins, Jonathan Wells, Joe Marsh, Cigall Kadoch. Fitness data all from The Cancer Dependency Map Cancer Data Science x.com/kadochlab/stat…

On the cover of Cell Systems: inspired by the classic album cover of Joy Division's Unknown Pleasures using fitness responses of 342 cancer cell lines to gene knockouts. Check out this excellent work by Joshua Pan and robin cell.com/cell-systems/f…


Selected for the cover of this month's Cell Systems issue is a design that Joshua Pan and I put together using #Rstats and @ClausWilke's ggridges package. cell.com/cell-systems/i…

The team behind The Cancer Dependency Map has worked hard to ensure quarterly public releases of newly generated data. This acknowledgement in @TraverHart lab's excellent preprint biorxiv.org/content/early/… is one example indicating a big payoff to #openscience. cc: Cancer Data Science Jesse Boehm
