
Hung Nguyen
@tienhung_91
Assistant Prof @UBChemistry | PD @UTChemistry | Computational modeling of RNA structure and folding, intrinsically disordered proteins and phase separation
ID: 3977449288
http://BioSimLabUB.github.io 17-10-2015 05:52:12
87 Tweet
289 Takipçi
611 Takip Edilen




Everything ready for the CECAM/Psi-k workshop in Berlin, organized by Cecilia Clementi with a little help from Michele Ceriotti, Gábor Csányi and Lixin Sun. If you wanted to come and couldn't get a spot, you can follow on zoom. Link & schedule: sites.google.com/view/cecam-psi…




Close one page, open another. Privileged to work with Dave and amazing colleagues Thirumalai Lab @ UT Austin. Learnt a lot from you!!

We are hiring! Check out three open positions Chemistry Department apply.interfolio.com/130963 Physical/Analytical Chemistry - C&EN (Chemical & Engineering News) post at: chemistryjobs.acs.org/job/39218/tenu… Chemjobber


Interested in applying to #Chemistry grad study? Learn about what UB Dept of Chemistry can offer in a virtual informational session. Ask us anything (including how to make such a cool poster)! Free to join and everyone is welcome! Pls RT. Register link buffalo.zoom.us/meeting/regist…


Happy to share our recent work on The Journal of Physical Chemistry Lett (doi.org/10.1021/acs.jp…). Thanks to Dave Thirumalai, Hung Nguyen, Naoto Hori and Thirumalai Lab @ UT Austin. Using coarse-grained MD simulations, we studied salt-dependent self-association of CAG repeat RNA sequences.




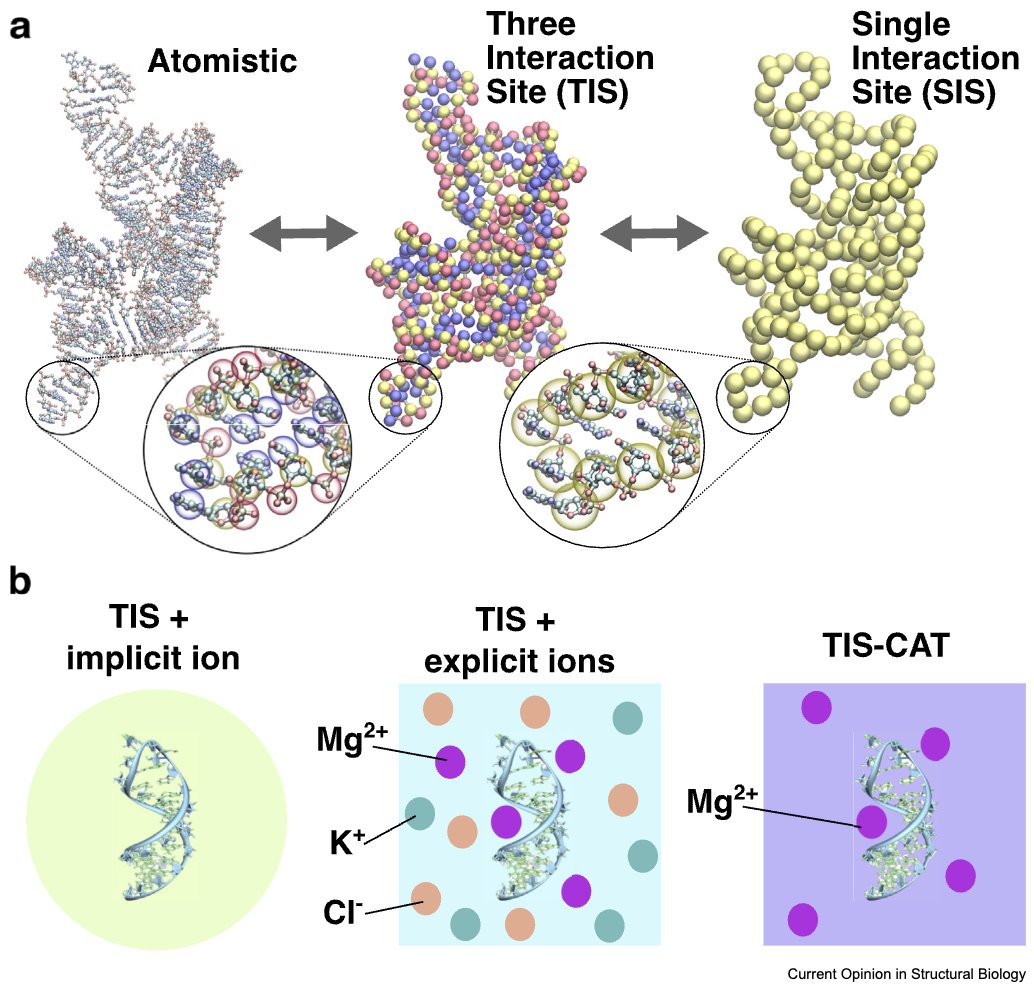
With Thirumalai Lab @ UT Austin and Hung Nguyen, we wrote a review on 'Minimal Models for RNA Simulations', now published in Current Opinion/Research in Structural Biology. We highlight how coarse-grained models capture the complex behaviour of RNA, from ribozyme folding to phase separation and more. doi.org/10.1016/j.sbi.…




![Sucheol Shin (@sucheol_shin) on Twitter photo RT plz! APS Focus Session "Genome Organization & Subnuclear Phenomena" is coming back at the 2025 Global Physics Summit! We invite the abstracts of latest developments in the 4D genome using experimental, theoretical, and/or computational methods, from all-level scientists! [1/4] RT plz! APS Focus Session "Genome Organization & Subnuclear Phenomena" is coming back at the 2025 Global Physics Summit! We invite the abstracts of latest developments in the 4D genome using experimental, theoretical, and/or computational methods, from all-level scientists! [1/4]](https://pbs.twimg.com/media/GZnz4xaWgAU7dO9.jpg)