
Yousuf A. Khan
@theyousufkhan
Scientist @Stanford. CryoEM/ET, AAA+, RNA, Recoding, ML. Formerly: DeepMind AlphaFold,EvoscaleAI, Churchill Scholar@Cambridge_Uni & seen on @Netflix
ID: 1083072925295611906
https://www.ncbi.nlm.nih.gov/myncbi/1DaKepGFnknwBd/bibliography/public/ 09-01-2019 18:48:02
1,1K Tweet
596 Followers
450 Following



Our CSO, Prof. Pasha Baranov, discusses the limitations of RNA-Seq and the necessity of Ribo-seq for a complete understanding. 📹 Watch the full video on YouTube here: youtu.be/YxrLpdYEoJ8 ✉️ Contact us at [email protected] #HarnessingTranslatomics

Flash invariant point attention enables the generation of larger biomolecules. Olivia Viessmann
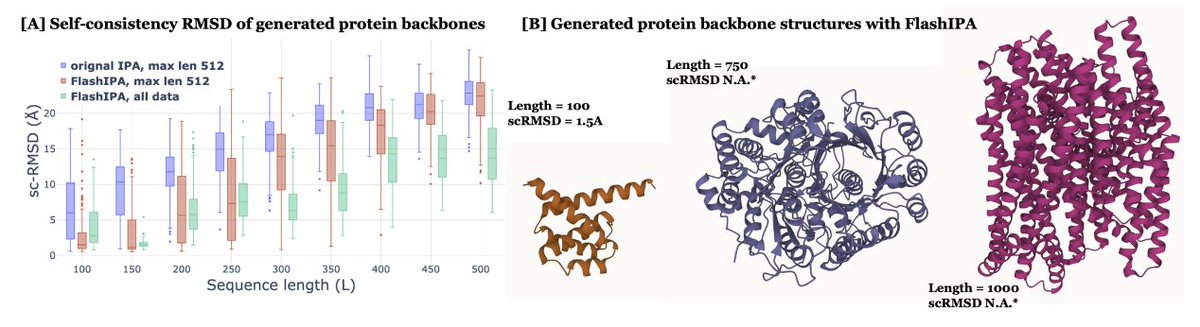







What would our data landscapes look like if we could biochemically characterize 100s, 1000s, 10^n evolutionary sequences? Papers like this make strides towards that moonshot dream!! Such neat work from Margaux Pinney, Ph.D. lab! science.org/doi/10.1126/sc…


Congratulations to Yousuf A. Khan and co-authors on the latest work (rdcu.be/euBhY), which reveals a unique “side loading” mechanism for Sec18/NSF-driven SNARE disassembly—offering fresh insight into membrane fusion! I’m proud to have collaborated on this work.






