Thomas Fleischer
@tf_82
ID: 826407701836087296
31-01-2017 12:32:04
32 Tweet
102 Takipçi
39 Takip Edilen


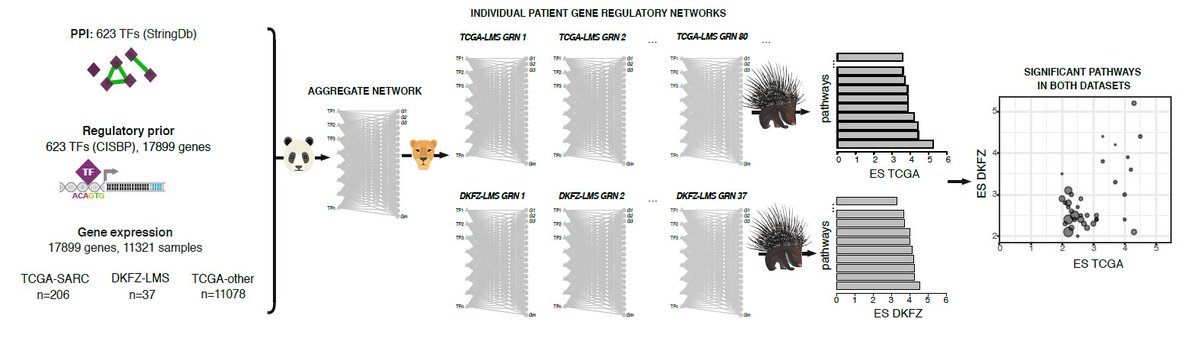
want to analyze patient-specific network models to detect heterogeneity? take a look at our tool PORCUPINE by Tatiana Belova kuijjerlab.org/tools/! a new version of the manuscript is now out on the BioRxiv doi.org/10.1101/2022.0…


Job opportunity: Postdoc position in computational biology. To be co-supervised by myself and Federico Gaiti. There is still time to apply. #epigenetics #Toronto #genomic #computationalscience #cancerevolution nature.com/naturecareers/…

Great collaboration with Valeria Vitelli, @ManuelaZucknick, @freeges, and Vessela Kristensen. Fun to be part of a project so heavily focused on Bayesian statistics!







Thank you Vessela Kristensen @freeges Alvaro Köhn-Luque Magnus Seierstad for a wonderful meeting in Oslo. See you next year in....to be announced!



🧬Opening for a postdoc in my group @NCMMnews Universitetet i Oslo UiO - Det medisinske fakultet @unioslo_mn to generate deep learning models to enhance JASPAR JASPAR database & UniBind, boosting gene regulation research. Project in collaboration with Anshul Kundaje (anshulkundaje@bluesky), Wyeth Wasserman. Plz share jobbnorge.no/en/available-j…





Please RT! 🧬🧬 I am looking for a #postdoc fellow to lead a new project in 3D chromatin organisation and transcriptional regulation in cancer (experimental and/or computational) at South Australian immunoGENomics Cancer Institute in Adelaide. Apply below: careers.adelaide.edu.au/cw/en/job/5146…

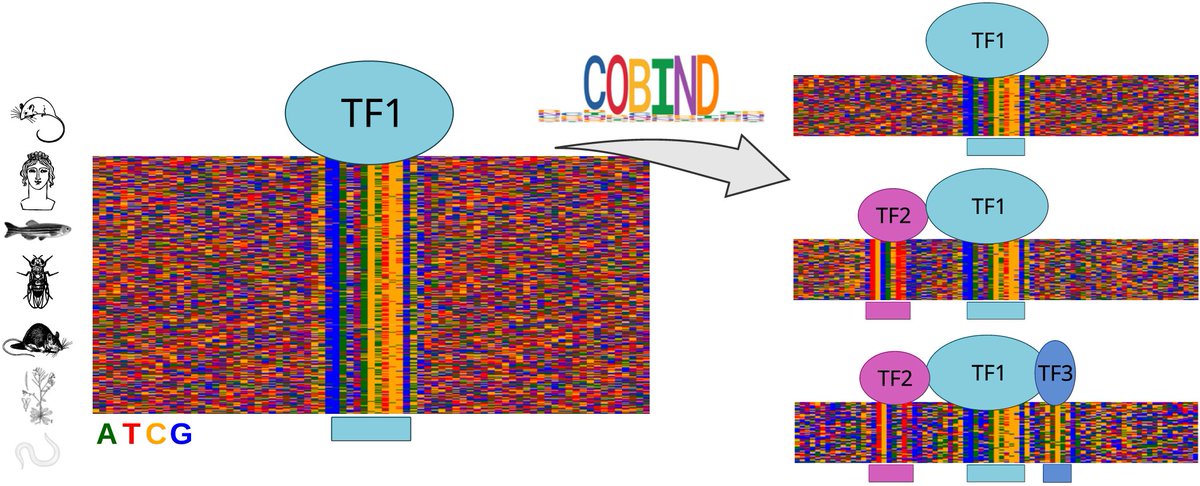
🎉 Excited to share the peer-reviewed version of our manuscript describing COBIND, a tool for revealing TF co-binding patterns using non-negative matrix factorization. Great work led by Ieva Rauluševičiūtė @NCMMnews Nordic EMBL P'ship UiO - Det medisinske fakultet Universitetet i Oslo academic.oup.com/nar/advance-ar… Nucleic Acids Res