
Jimin Tan
@tan_jimin
Postdoc at NYU Langone (ML/Genomics/Imaging)
ID: 1860923389
http://tanjimin.com 13-09-2013 15:11:43
19 Tweet
85 Followers
89 Following

this is awesome! great job, team! i just wanted to plug in a project from the capstone project last year at NYU Center for Data Science done by a team of master's students (Jimin Tan, Chenqin Yang, Yakun Wang, Yash Deshpande) which was on unsupervised video dubbing tanjimin.github.io/unsupervised-v…

Very excited to present our latest work with Jimin Tan and Bo Xia: using machine learning to predict cell type-specific chromatin architecture and enable in silico perturbation studies! NYU Grossman School of Medicine Perlmutter Cancer Center at NYU Langone Health biorxiv.org/content/10.110…
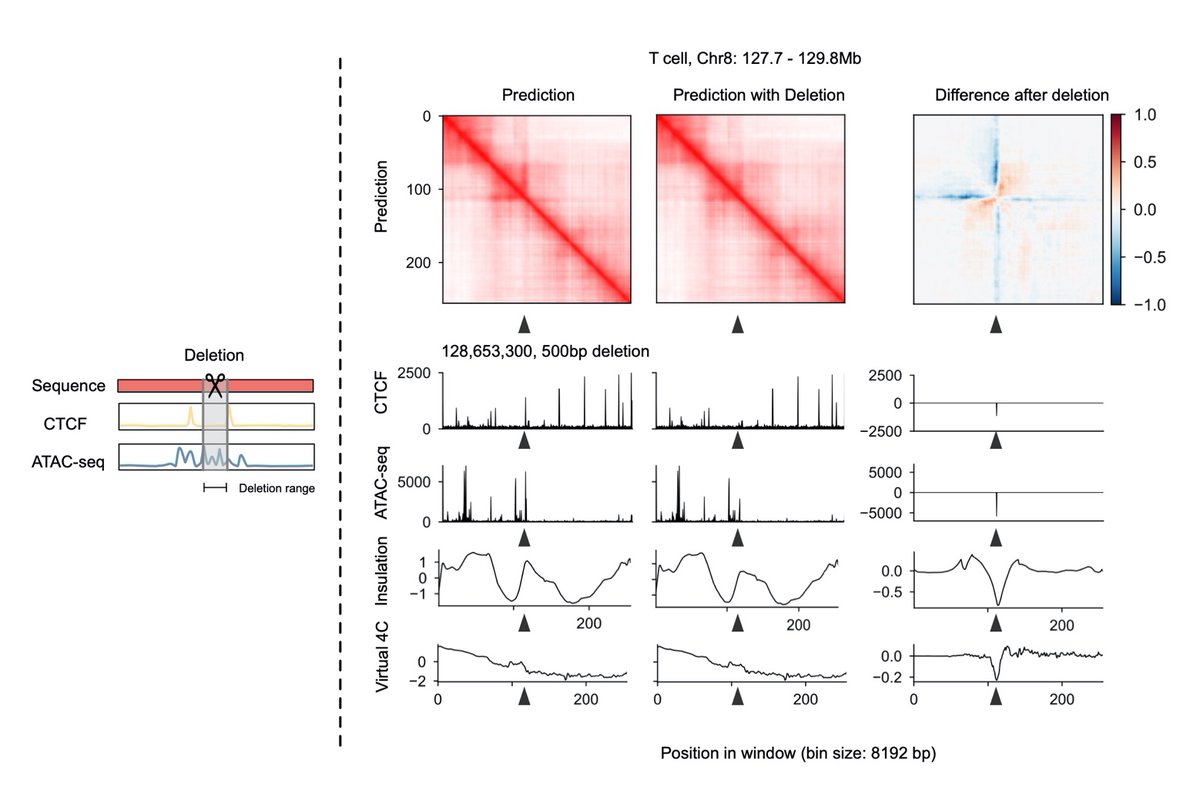



Our C.Origami work is online Nature Biotechnology! A tour de force by Jimin Tan and a wonderful collaboration with Aristotelis Tsirigos. Check out how we use high-throughput in silico genetic screen to discover new regulation of genome organization. Full manuscript here: rdcu.be/c23mx

C.Origami, a machine learning model for cell type-specific chromatin architecture prediction and discovery of putative regulatory elements! Developed by our student Jimin Tan in collaboration with Bo Xia. Nature Biotechnology 4DNucleome EpigeneticsChromatin epigenetics_papers





Former CDS MS student Jimin Tan, CDS Professor Kyunghyun Cho, & others have developed an AI model that identifies tumor microenvironments. Their work, published in Nature Biomedical Engineering, uncovered a new lung cancer subtype linked to poor survival. nyudatascience.medium.com/ai-model-disco…


Thrilled to share that I’m joining MGH Pathology, Harvard Medical School & Broad Institute as an Assistant Professor! 🚀 The Xia Lab is entering a new chapter at both MGH and Broad - building biologically informed ML/AI models to decode genome regulation & engineer cell fates. Stay tuned




