Mara Stadler
@stadlermara
👩🏼💻PhD student in Stats @BioDataSc @MunichDS, interested in high-dimensional stats & modeling combinatorial effects 📊📈 & bio application 🧬🦠
ID: 1204779027866816512
https://marastadler.github.io/mara-stadler/ 11-12-2019 15:04:54
37 Tweet
98 Followers
177 Following


📊 Unveiling Stable Interactions in High-Throughput data! 💡Mara Stadler introduce #robust #statistical workflow: 🔗 Application to interaction effects between chromatin modifications 🧬 Lasso model for hierarchical interactions 🔎 Stability-based model selection #ISMBECCB2023


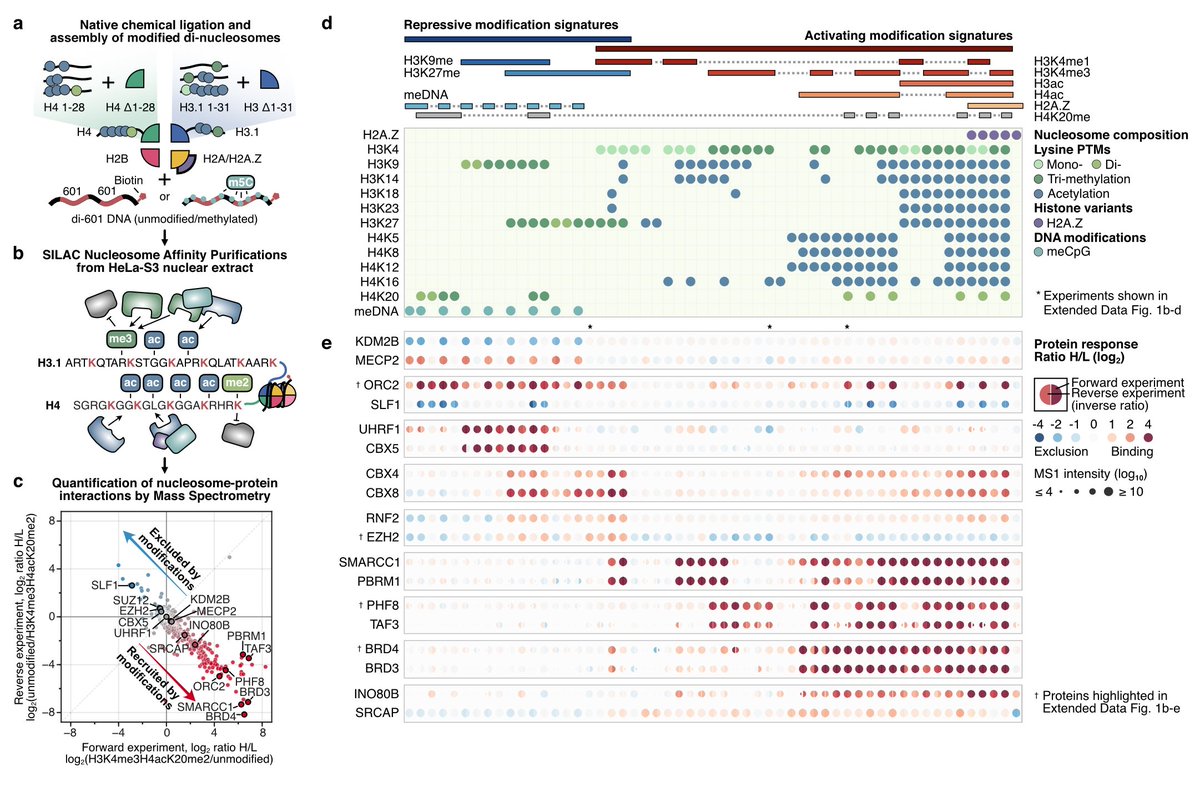
Publication alert ! Paper from helmholtz-munich.de/en/ife in nature How complex epigenetic modification signatures regulate genome function. With interactive online resource marcs.helmholtz-munich.de Big data ! Epigenetics Research at Helmholtz Munich Helmholtz Munich | @HelmholtzMunich nature.com/articles/s4158…



Check out this amazing data resource to study how chromatin modifications regulate protein binding behavior by Bartke lab and Robert Schneider 🧬 I'm very proud to be part of this project 👇