Sairam Behera
@srbehera11
Research fellow at @BCM_HGSC
ID: 762116449
https://srbehera.github.io/ 16-08-2012 18:50:39
2,2K Tweet
490 Takipçi
1,1K Takip Edilen

don't use hg38.fa as-is. checkout the references 😜 here: ftp-trace.ncbi.nlm.nih.gov/ReferenceSampl… rendered the ipynb (not mine) here: gist.github.com/brentp/1935e9b… in short, use: GRCh38_GIABv3_no_alt_analysis_set_maskedGRC_decoys_MAP2K3_KMT2C_KCNJ18.fasta.gz other updates on the best hg38 reference?


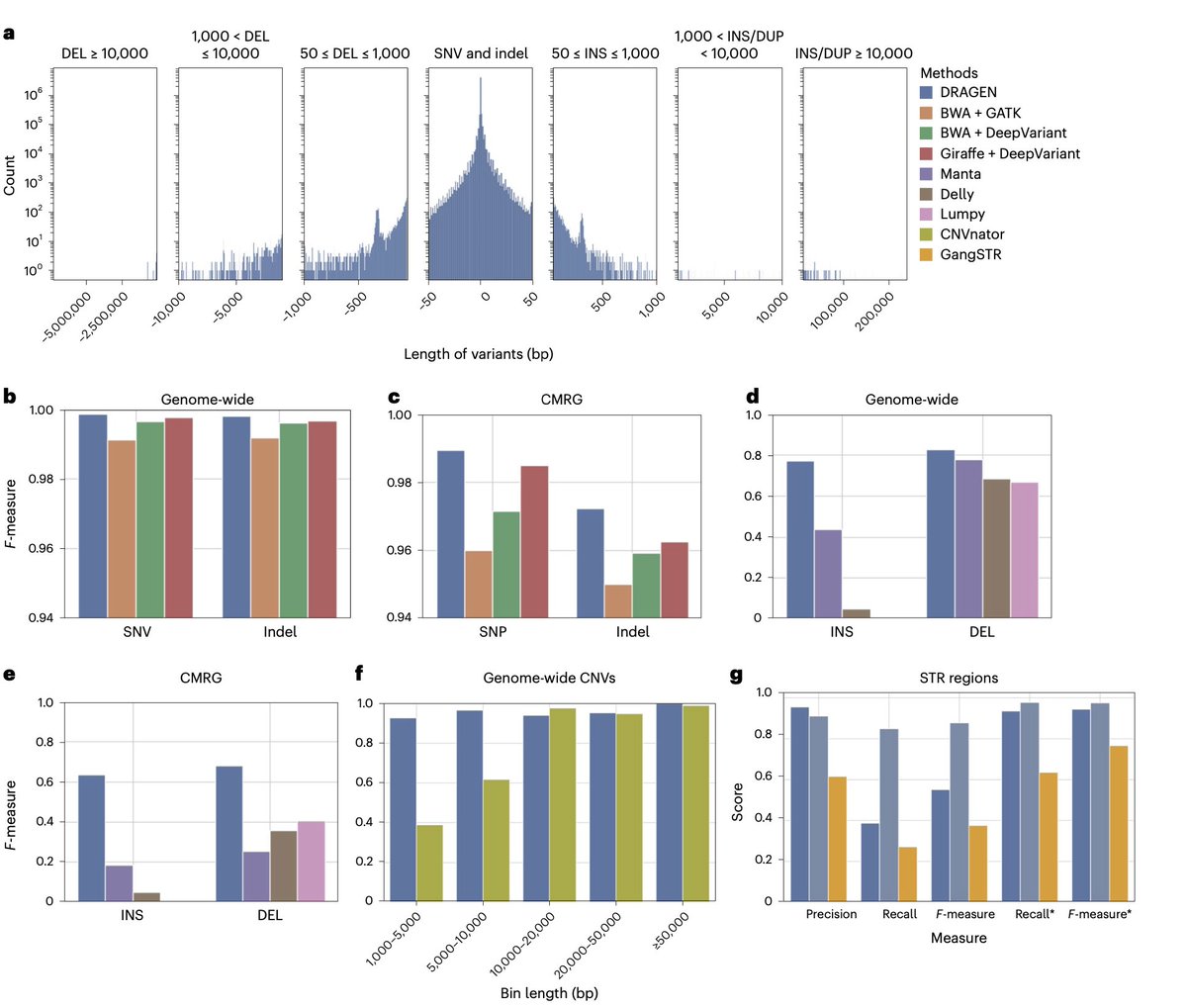
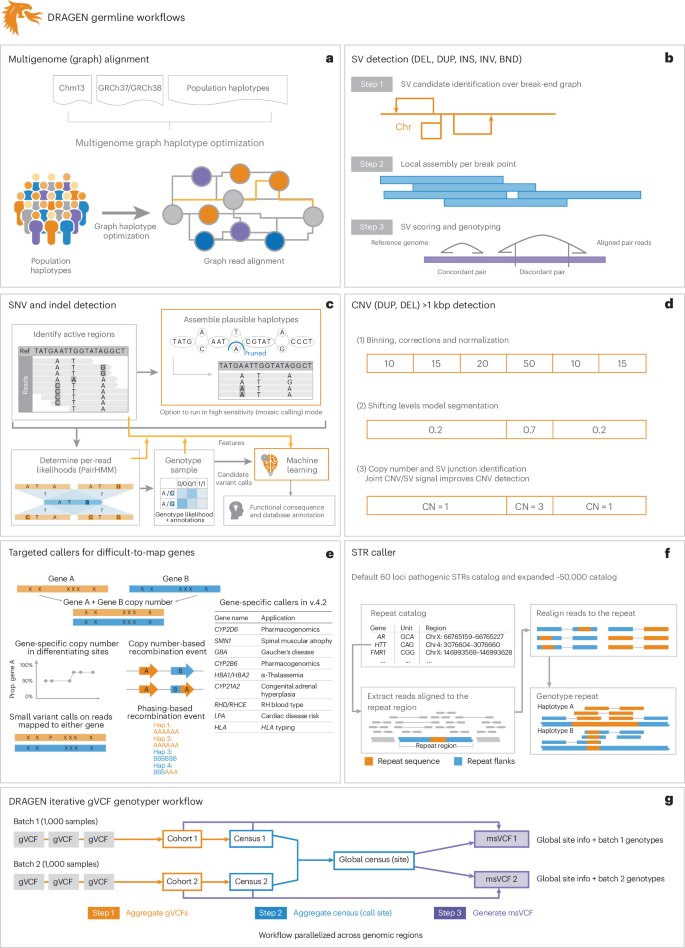
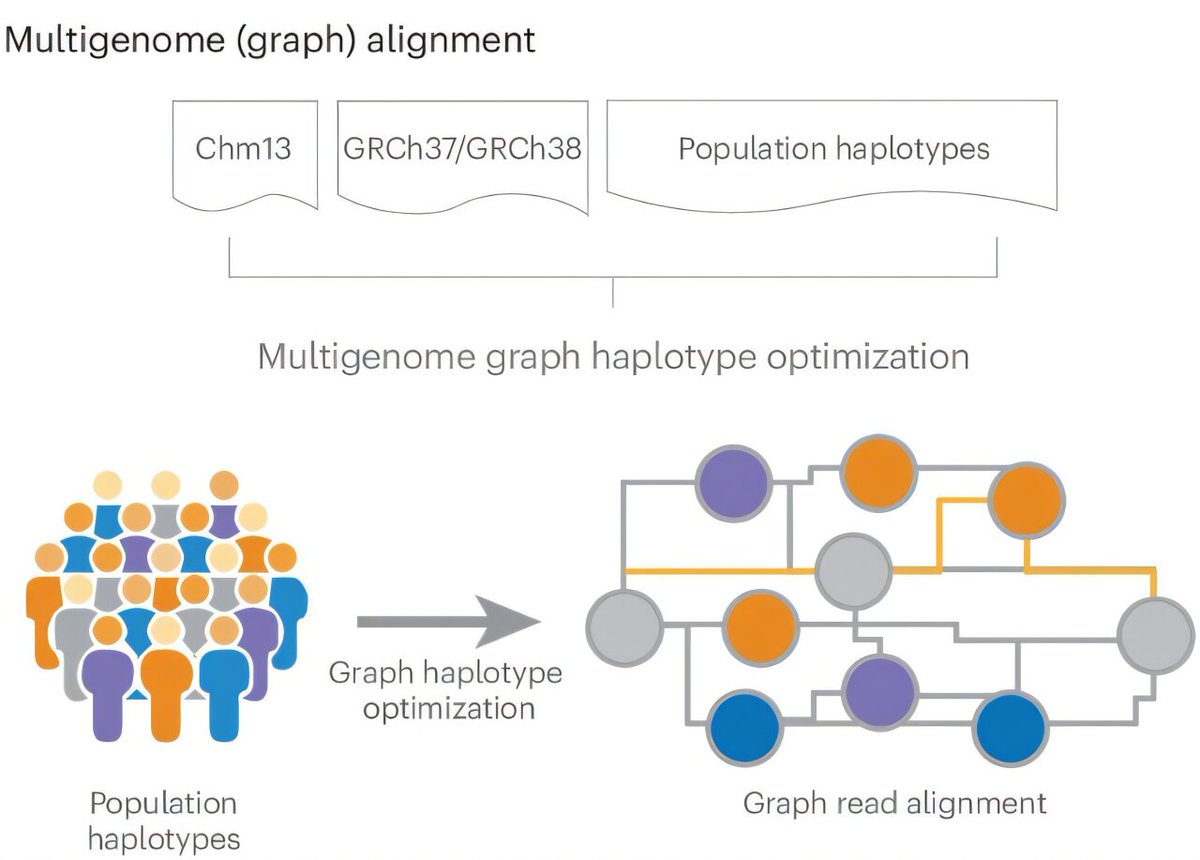
#DRAGEN Illumina method fully described & benchmarked out in Nature Biotechnology: nature.com/articles/s4158… Graph genome based detection of SNV, STR, SV, CNV all in 30 min! Improvements in all variants! Huge thanks to Severine Catreux Rami mehio Sairam Behera BCM HGSC #Bioinformatics 1/5


#DRAGEN is now out in Nature Biotechnology . We outline the method, which has been benchmarked and also analyzed using the 1KGP cohort. It demonstrates accuracy and scalability through graph genome and machine learning approaches. nature.com/articles/s4158…


New in Nature Biotechnology: Illumina #DRAGEN marks milestone in sequencing data analysis, enabling insights across various diseases with a highly comprehensive, scalable #bioinformatics platform. Sairam Behera Severine Catreux Rami mehio Fritz Sedlazeck nature.com/articles/s4158…


Fritz Sedlazeck Rami mehio Severine Catreux Sairam Behera BCM HGSC Illumina BCM Department of Molecular and Human Genetics BCMHouston Rice Computer Science Validation testing of next-gen genome analysis platform reveals potentially disruptive tech #NBTintheNews via Phys.org phys.org/news/2024-11-v…


The Long-read sequencing Special Issue is now LIVE! Guest-edited by Alexander Hoischen, Fritz Sedlazeck, and AnaConesa!

Our first curated draft somatic structural variant benchmark for the new GIAB PDAC tumor cell line HG008-T is at ftp-trace.ncbi.nlm.nih.gov/ReferenceSampl…, based on extensive short+long read sequencing data described in doi.org/10.1101/2024.0…. Feedback to improve future versions is very welcome!

The new issue of Genome Research is now live! Follow the link to new research on mitochondrial–nuclear gene expression regulation, guppy Y Chromosome genomics, and more! tinyurl.com/Genome-Res-35-3


We summarized state-of-the-art computational methods for DNA methylation analysis using long-read sequencing, covering everything from base calling to sample-level, cell-type-level, and even population-scale analysis. Huge Thanks to Fritz Sedlazeck and Winston Timp for this great work!