Sophia Vincoff
@sophievincoff
AI+Protein Design 💻🧫 PhD student in Chatterjee Lab @DukeU
ID: 1676627288018898945
05-07-2023 16:29:04
11 Tweet
103 Followers
139 Following

Check out our lab's perspective on generative design of therapeutic binders published in COBME: sciencedirect.com/science/articl…! 💻🧫 Great work from my PhD students: Leo Chen Lauren Hong Vivian Sophia Vincoff Feel free to cite the review in your next manuscript! 😊#GenerativeAI

SaLT&PepPr is published in Communications Biology! 🥳 Here, we fine-tune the ESM-2 pLM to identify peptidic binding sites on target-interacting partner sequences. We fuse these "guide" peptides to E3 ubiquitin ligases to degrade disease-causing proteins! 💻➡️🧫 (1/n) nature.com/articles/s4200…


Super excited to share our new protein language model: FusOn-pLM, developed by my wonderful first-year PhD student Sophia Vincoff and her team of amazing undergrads!! 🎉 Sure, we know AlphaFold3/RFDiffusion and other structure prediction/design methods are great -- but what


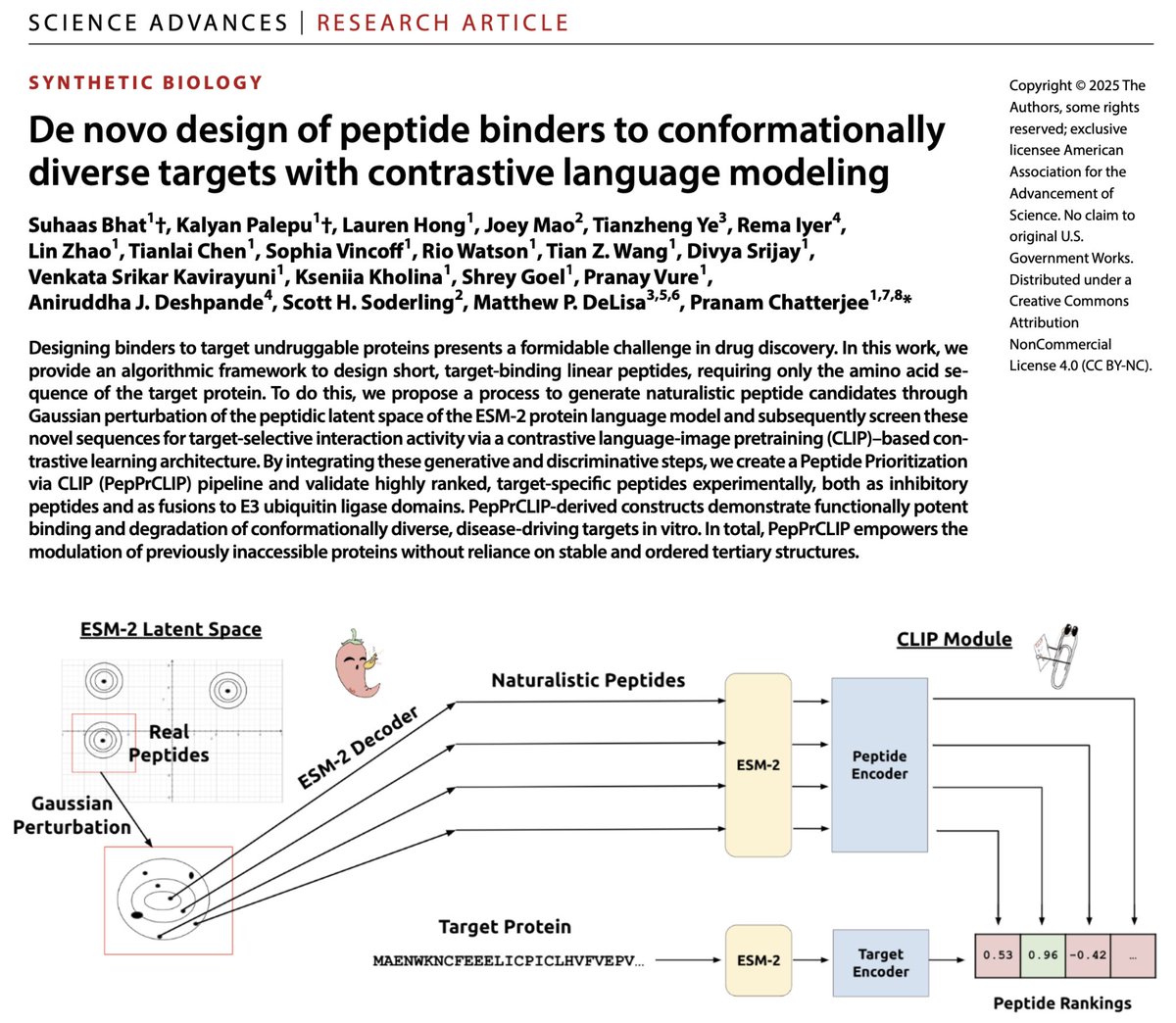
I'm usually not too emotional about paper acceptances, but this one justifies it. 🥹 It doesn't feel real, but PepPrCLIP is now published at Science Advances! Let me tell you its story. 📕 📜: science.org/doi/10.1126/sc… 💻: huggingface.co/ubiquitx/peppr… PepPrCLIP (🌶️📎) began as

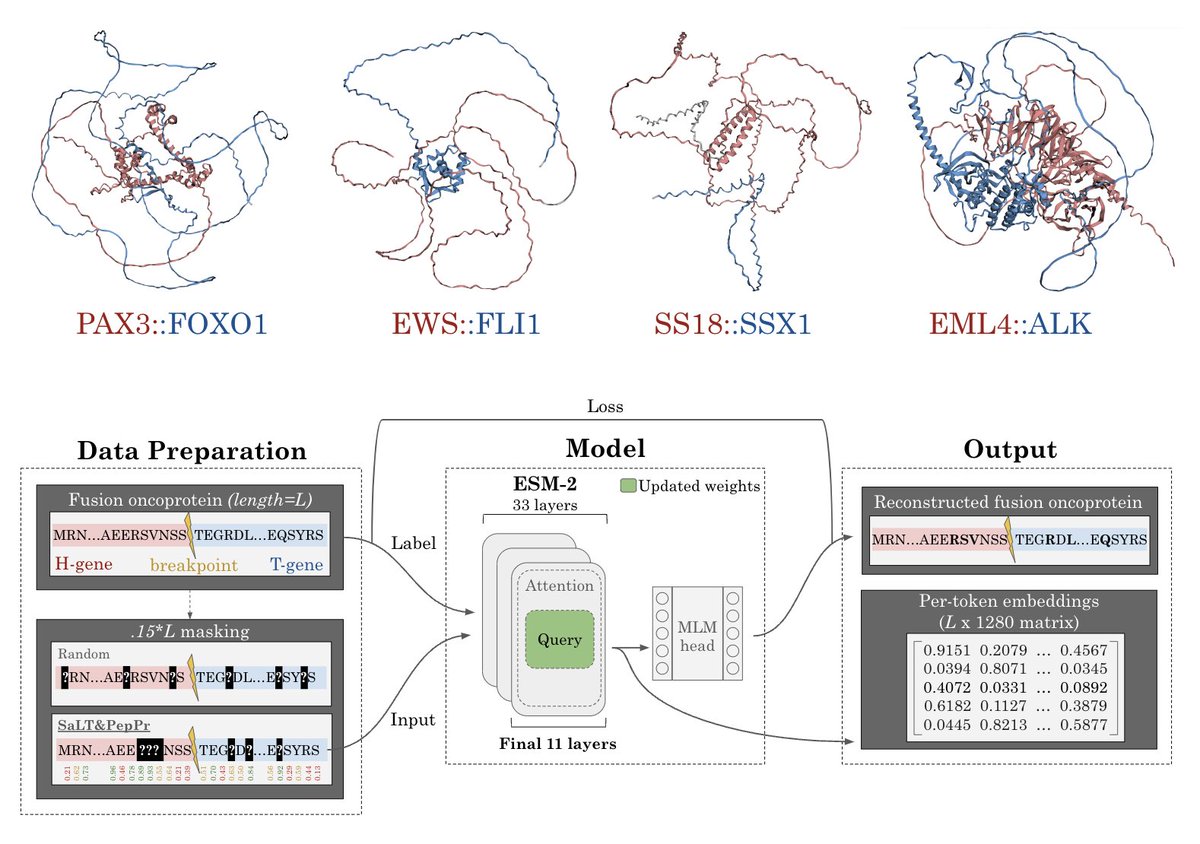
I'm excited to share my first published project as a PhD student! I want to thank my incredible advisor Pranam Chatterjee, brilliant undergrads Shrey Goel, Kseniia Kholina, Rishab Pulugurta, and Pranay Vure for their work, and the amazing Lauren Hong for drawing FusOn-pLM's logo.

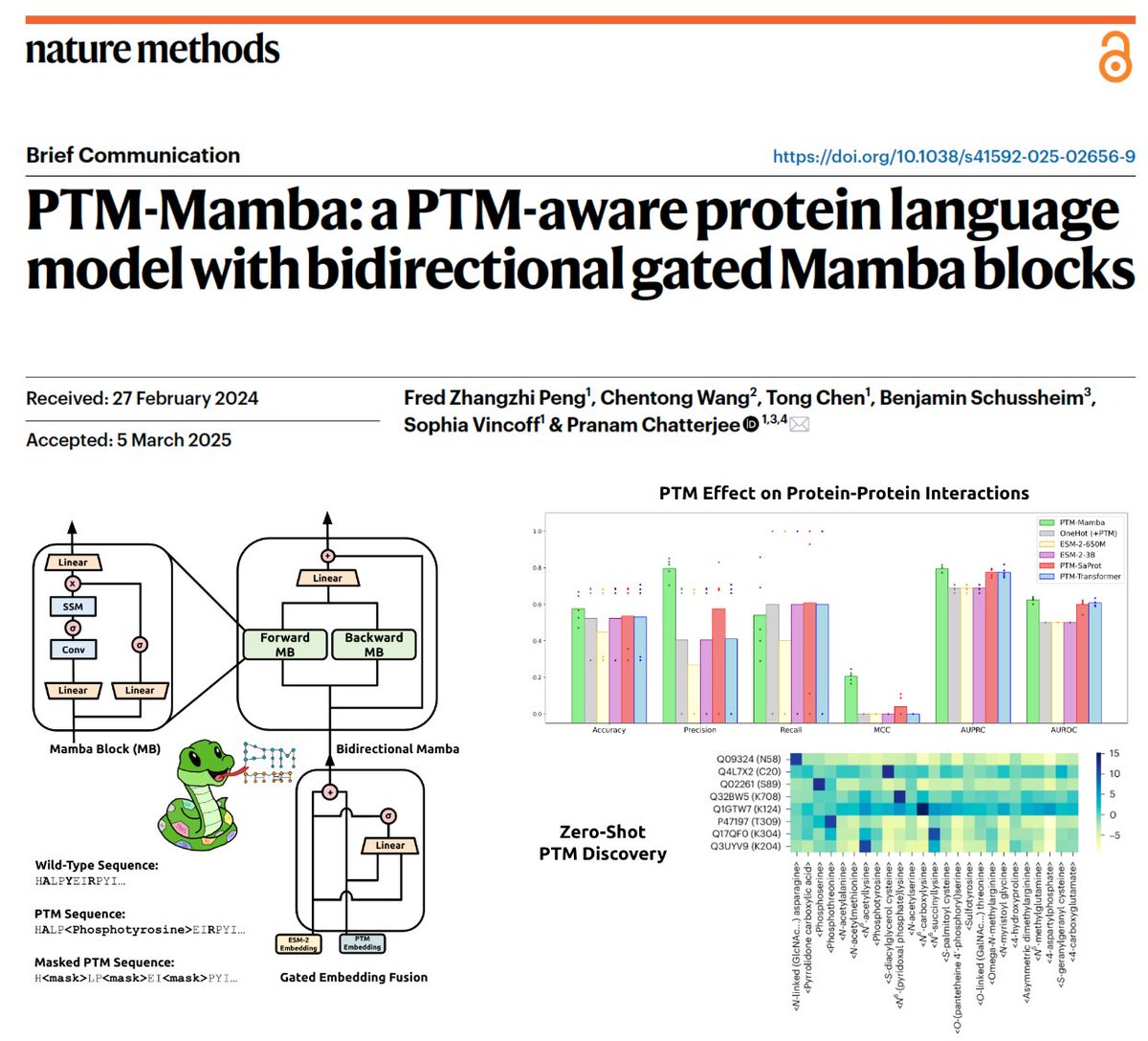
PTM-Mamba is out in Nature Methods!! 🥳 PTM-Mamba is the first/only pLM to encode both WT and post-translationally modified residues as tokens! And we do this without losing any base protein information from ESM-2! 🤝 📜: nature.com/articles/s4159… 💻: github.com/programmablebi… We

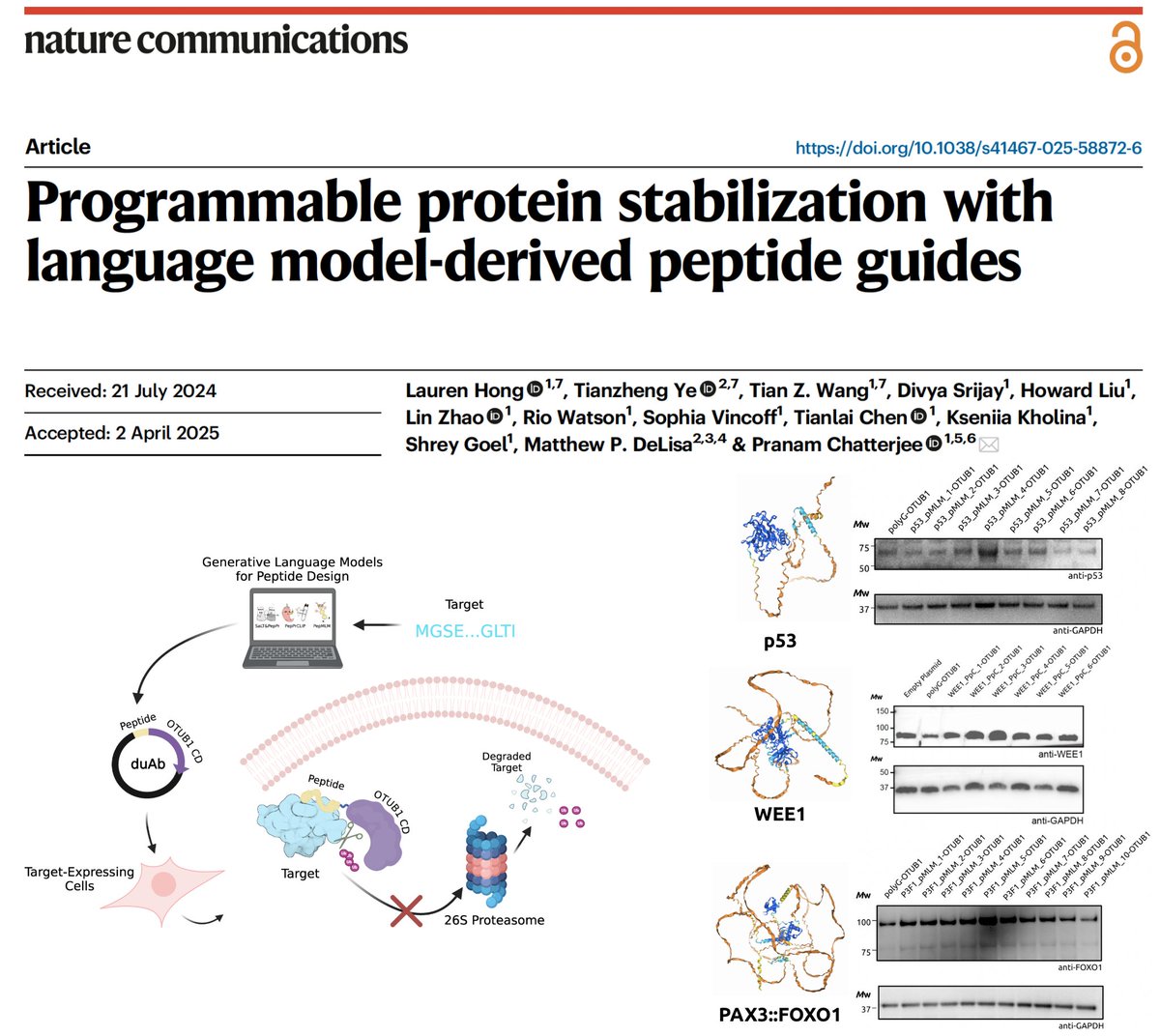
Our pLM-designed peptides, when attached to E3 ligases (aka uAbs), enable us to programmably degrade disease targets (think CRISPRi, but for proteins). But what if we want to STABILIZE useful proteins? In Nature Communications, we introduce duAbs! 🌟 📜: nature.com/articles/s4146… 🧬: