Simon Mathis
@simmat20
PhD @Cambridge_Uni | AI in protein design & engineering | biotech & environmental applications | enzymes | geometric DL | prev. @ETH_physics
ID: 1255778628614475777
http://simonmathis.org 30-04-2020 08:39:13
205 Tweet
909 Takipçi
953 Takip Edilen





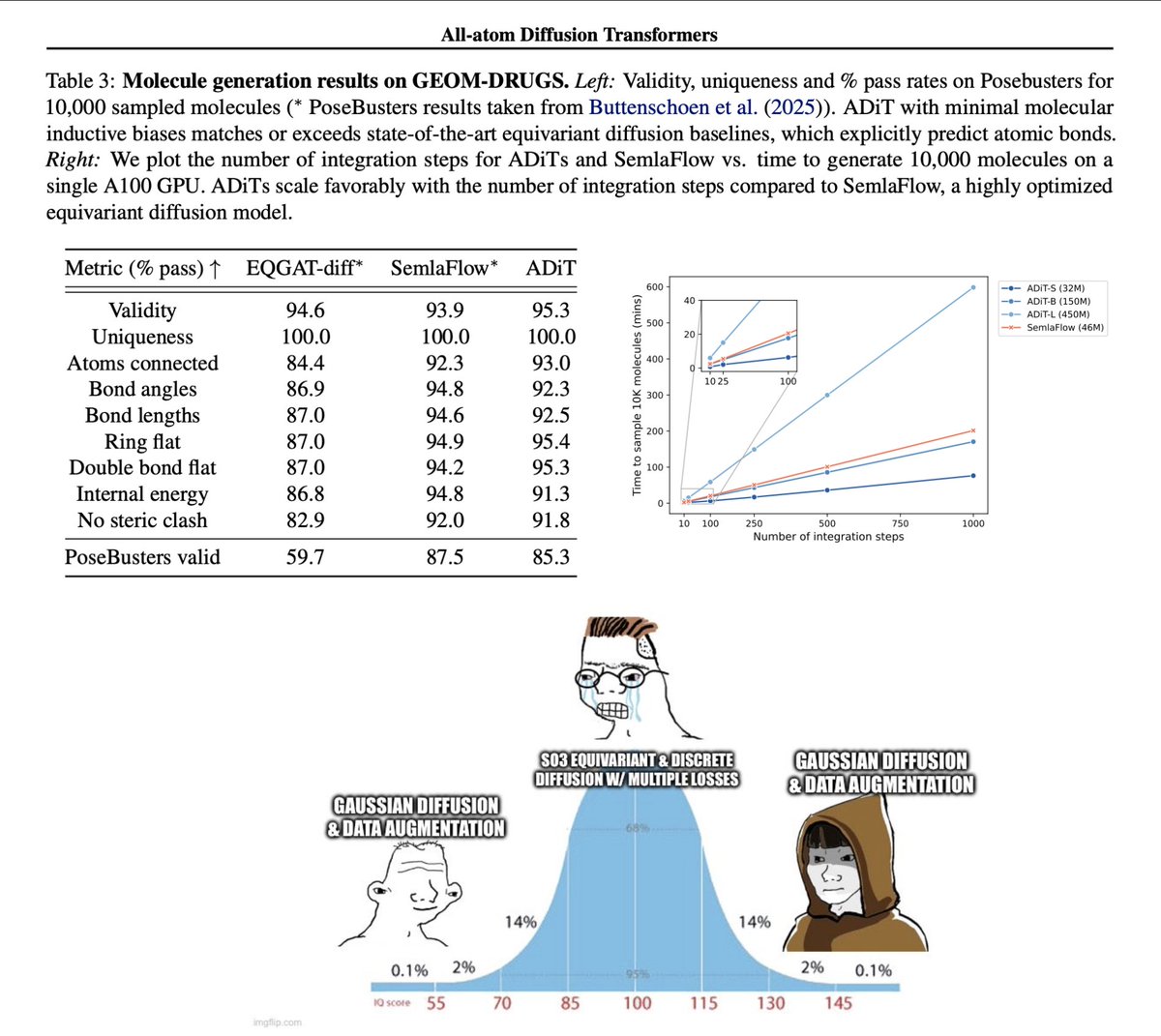
Check out La-Proteina, our new model for all-atom structure generation at scale! Was a very fun project to work on with Tomas Geffner and the rest of the team. Two things I am particularly excited about in 🧵 1/n


RNA-FrameFlow has found a new home in TMLR Accepted papers at TMLR 🥳 Over the past one year, we've spent quite some time making our method faster, developing more intricate evaluation pipelines, thinking about conditional design, and assessing/ablating our modeling choices Here are some

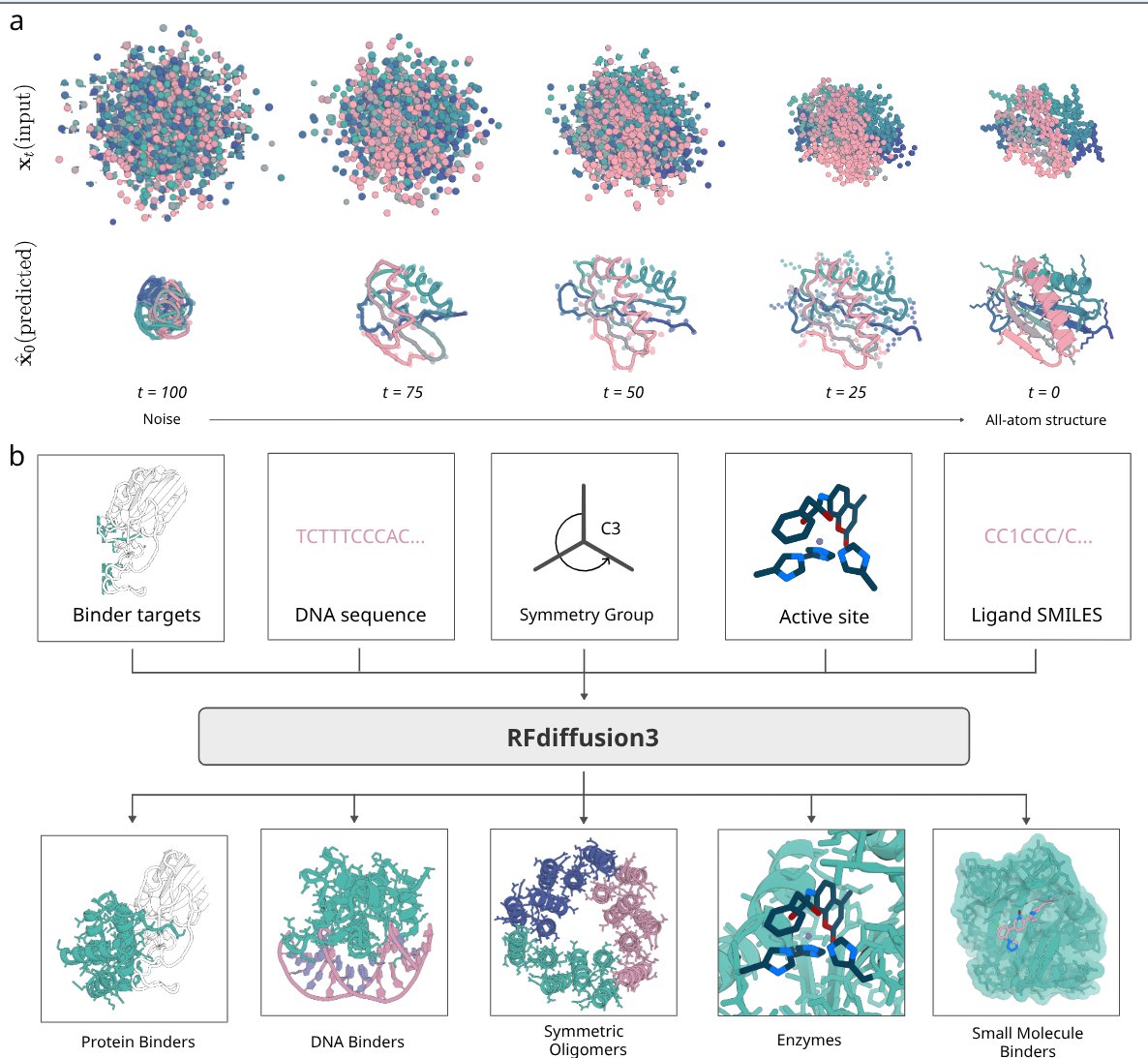
RosettaFold 3 is here! 🧬🚀 AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release! Congratulations Simon Mathis and team!!! ❤️



Atomsworks and RF3 is finally here!! I was very fortunate to get early access to atomworks and it really lowers the activation energy todo some really cool stuff with the PDB very quickly. Might share fun examples later Huge congrats to Simon Mathis and team!!! ✨


Great to finally have this available for people 🙌 Thank you to everyone involved, especially Nate Corley & Rohith Krishna! We'll be pushing a few smaller updates in the coming days to enhance usability for everyone (including some docs on what you can do with it) - so stay tuned (:

Where are all the de novo designed therapeutics? In a guest post by Frédéric Dreyer, we break down what the actual bottlenecks, challenges, and opportunities are in building and deploying purely in silico drug discovery engines, with a focus on antibody structure. Link below 👇


Check out our new perspective "Illuminating the universe of enzyme catalysis in the era of artificial intelligence" now out in Cell Systems ! We discuss a vision and path forward for genetically encoding almost all chemistry, powered by new AI tools: authors.elsevier.com/c/1lg4h8YyDfuZ…