
Sheridan Cavalier
@sheridancavali1
@BCMB_JHMI PhD candidate at #johnshopkins studying synaptic formation of memory
Full-time Texan, part-time scientist
Email: [email protected]
ID: 1313900773718192128
07-10-2020 17:55:37
20 Tweet
54 Followers
60 Following

Our grad students deserve better, Johns Hopkins University. They could use a bonus right now. #priorities

Ariel Gershman with a great 2 minute flash talk on her work looking at epigenetics on a complete human genome #AGBT22 #T2T


At #AGBT, Sheridan Cavalier will present their team's research: 'Single-cell transcript #isoform sequencing of the actived adult mouse hippocampus with 10x #Genomics and Oxford #Nanopore'. Find out more: bit.ly/3xc4EjY #AGBT22


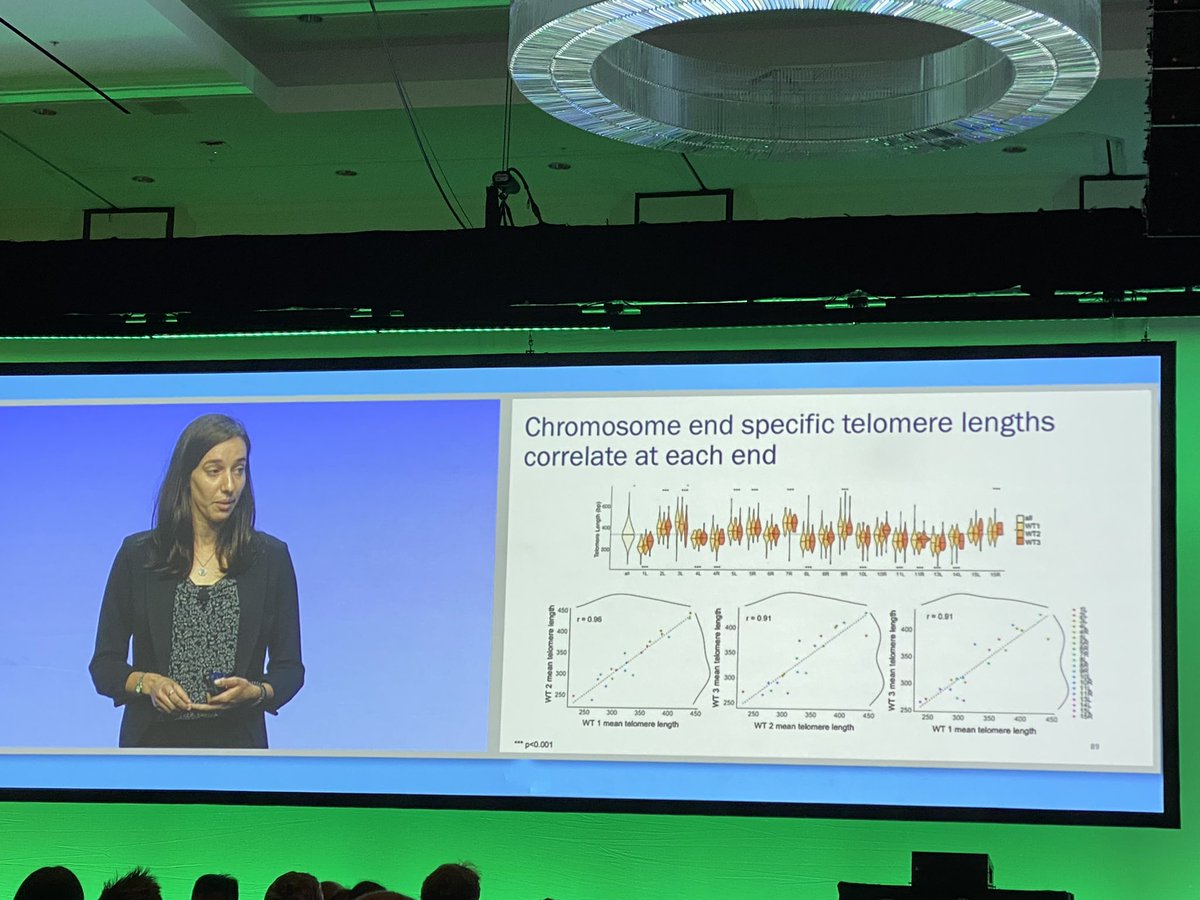
Long native Oxford Nanopore reads used to measure telomere length in yeast and reveals significant biological variation #AGBT22

After dinner, check out Sheridan Cavalier, PhiD 's talk in the Technology Session (7:50PM) on "Single-cell transcript isoform sequencing of the activated adult mouse hippocampus with 10x Genomics and Oxford Nanopore"


#AGBT22 Cavalier, John Hopkins Neuroscience. Activity induced transcription produces alternative splicing at single cell in brains by 10x Genomics and Oxford Nanopore . Used R2C2, using RCA, then concatamer on ONT for alt splicing.
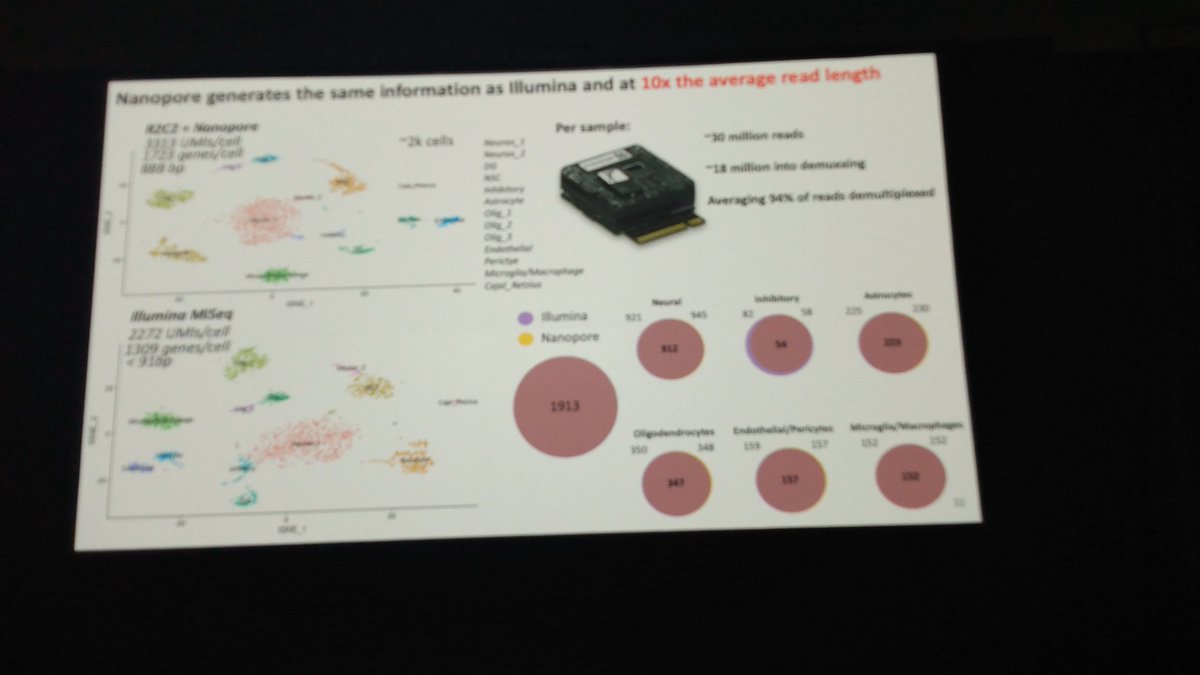

.Sheridan Cavalier, PhiD from Winston Timp lab: identifying and quantifying isoforms is easy using Oxford Nanopore full length RNAseq and the fragment length distribution we see on the tape station is the same as the resulting read length #AGBT22







So happy we were all binned together @binningsingletons.com on bluesky ♥️🦠




