Bin Shao
@shaobin_phy
Broadie. deep learning; synthetic biology; single cell genomics; non-linear dynamics. opinions are my own.
ID: 807050606078820354
https://www.biorxiv.org/content/10.1101/2024.12.30.630741v2 09-12-2016 02:33:53
507 Tweet
711 Followers
1,1K Following









Thrilled that our work on this problem with Karel Břinda, Zamin Iqbal, and others is out in Nature Methods today! We used phylogenetic compression (described in the thread) to compress every microbe ever sequenced onto a flash drive so that it can be searched with a laptop!

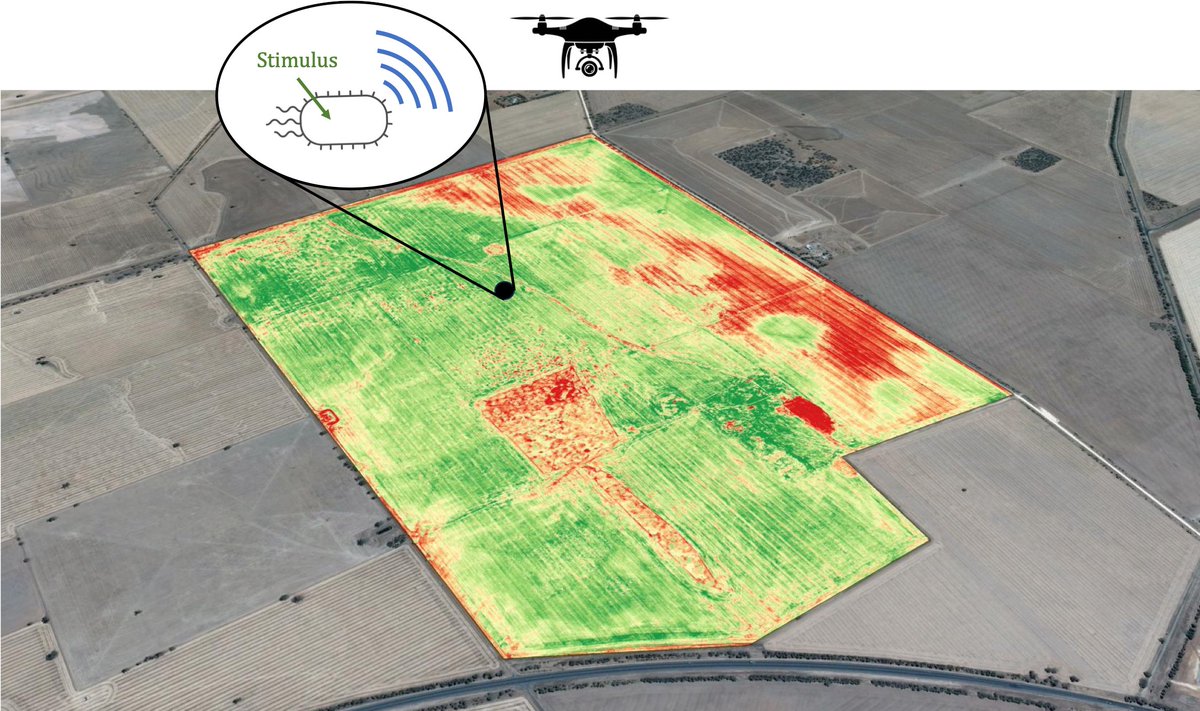
Seeing what bacteria see from a flying UAV 👁️ rdcu.be/ehkQB #synbio #biotech Itai Levin Yonatan Chemla