Shailesh Kumar
@shaileshgenome
Scientist at National Institute of Plant Genome Research (NIPGR). Bioinformatics, Genomics, Big data analysis, Plant Biotechnology.
ID: 372717557
http://www.nipgr.res.in/research/dr_shailesh.php 13-09-2011 08:54:24
39 Tweet
253 Takipçi
450 Takip Edilen

It was an absolute honour to present at Embassy of India in Japan India in Japanインド大使館 in 2018. Great interaction with scientists from different institutions at Tokyo, Japan.


📌Prof. K. Thangaraj speaks on '𝑷𝒐𝒑𝒖𝒍𝒂𝒕𝒊𝒐𝒏 𝑮𝒆𝒏𝒐𝒎𝒊𝒄𝒔 𝒂𝒏𝒅 𝑷𝒖𝒃𝒍𝒊𝒄 𝑯𝒆𝒂𝒍𝒕𝒉' in our new seminar series "𝑴𝒆𝒄𝒉𝒂𝒏𝒊𝒔𝒎𝒔 𝒕𝒐 𝑴𝒆𝒅𝒊𝒄𝒊𝒏𝒆" 📆: Aug 26, 2021 | 4.00PM Department of Biotechnology Dr. Renu Swarup @DBT_CDFD Bangalore Life Science Cluster (BLiSC) SciCommIndia 🇮🇳 #inStemTalkSeries





Dr. Shailesh Kumar's team at NIPGR has developed a computational pipeline for the identification of tRNA-derived small ncRNAs (tncRNAs) in small RNA sequencing (sRNA-Seq) datasets, & studied various features of these novel molecules. Read more: doi.org/10.1016/j.csbj… Department of Biotechnology
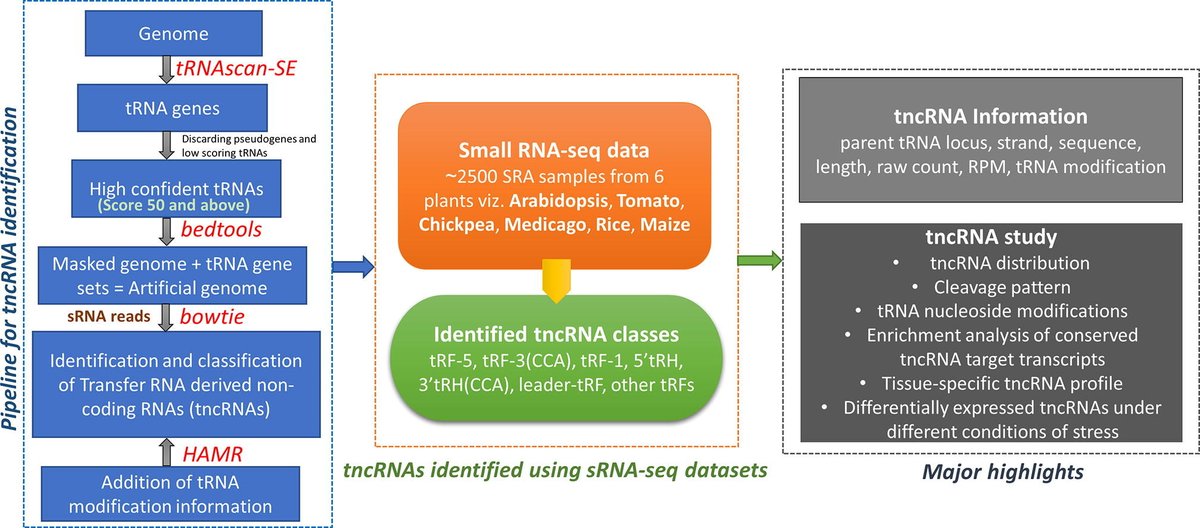

The workflow, software package, and results are freely available at nipgr.ac.in/tncRNA. Shailesh Kumar Department of Biotechnology SciCommIndia 🇮🇳 CSBJ


Dr. Shailesh Kumar's research group at NIPGR has developed a Machine Learning method for the prediction of Antimicrobial peptide prediction and designing of better therapeutic peptides. Server Link: nipgr.ac.in/PTPAMP Publication Link: link.springer.com/article/10.100… Department of Biotechnology

(1/2) Dr. Shailesh Kumar delivered a lecture on 'Role of next generation sequencing datasets in genomics research' for undergraduate students from Institute of Home Economics, University of Delhi on August 05, 2022 -- Department of Biotechnology SciCommIndia 🇮🇳 SERB Shailesh Kumar


Delighted to share our article introducing a simple method for detecting rsRNAs in Arabidopsis thaliana (doi.org/10.1016/j.mex.…). Co-authored with outstanding lab mates:Garima Kalakoti Ajeet Singh Srija. Thanks to Shailesh Kumar for invaluable support. BRIC-NIPGR

Latest pipeline developed by our research group, for the identification of Small Open Reading Frames (smORFs) encoded plant-derived antimicrobial peptides smAMPsTK pubmed.ncbi.nlm.nih.gov/37464885/ BRIC-NIPGR Department of Biotechnology Rajesh Gokhale

Our latest research article on plant-based drug designing! In silico studies of alkaloids and their derivatives against N-acetyltransferase EIS protein from Mycobacterium tub… pubmed.ncbi.nlm.nih.gov/37728544/ BRIC-NIPGR Department of Biotechnology

We have developed a tool for rRNA fragment identification and characterization. Comprehensive profiling of rRNA-derived small RNAs in Arabidopsis thaliana using rsRNAfinder pipeline pubmed.ncbi.nlm.nih.gov/38089152/ nipgr.ac.in/rsRNAfinder Department of Biotechnology BRIC-NIPGR

In our recent article, we studied the Long non-coding RNA and microRNA landscape of two major domesticated cotton species ncbi.nlm.nih.gov/pmc/articles/P…… BRIC-NIPGR Department of Biotechnology

tncRNA Toolkit: A pipeline for convenient identification of RNA (tRNA)-derived non-coding RNAs pubmed.ncbi.nlm.nih.gov/36632599/ BRIC-NIPGR Department of Biotechnology

PTPAMP: prediction tool for plant-derived antimicrobial peptides pubmed.ncbi.nlm.nih.gov/35864258/ Department of Biotechnology BRIC-NIPGR

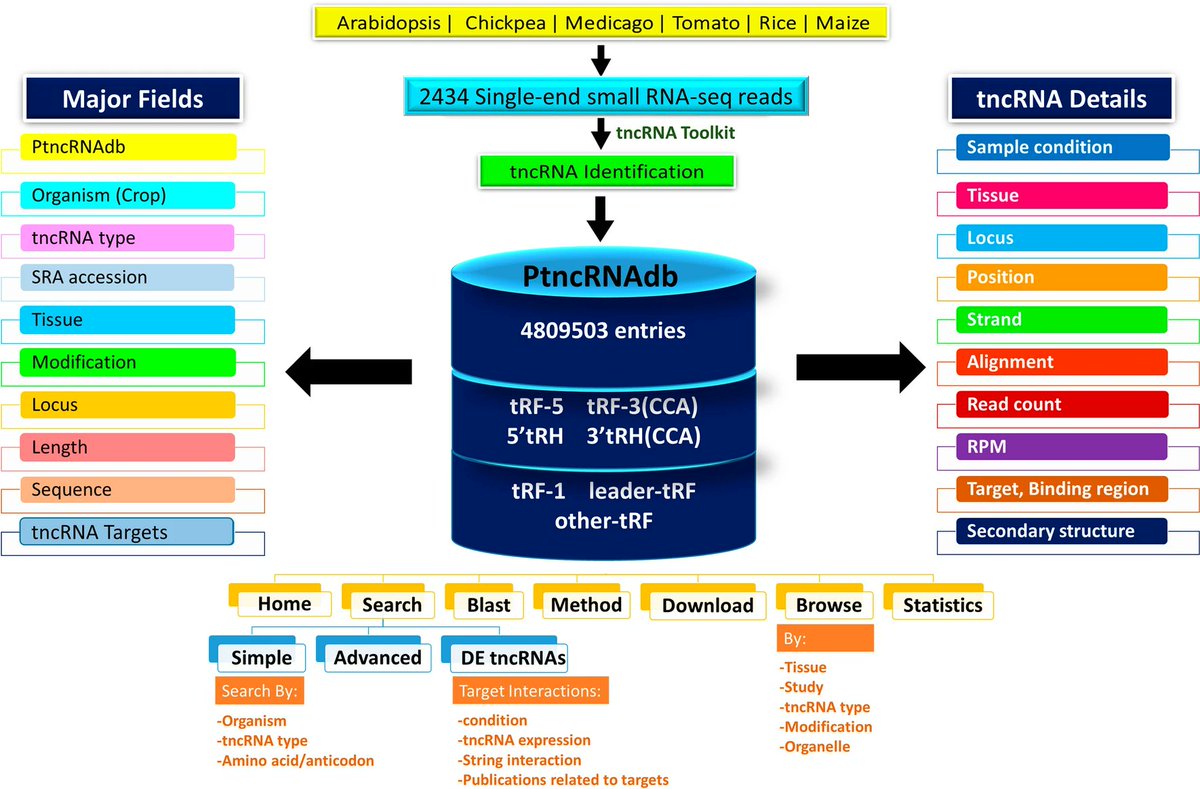
We have developed web resource for plant tRNAs. PtRNAdb: a web resource of plant tRNA genes from a wide range of plant species pubmed.ncbi.nlm.nih.gov/35875176/ nipgr.ac.in/PtncRNAdb BRIC-NIPGR Department of Biotechnology

New web resource from our research group !! PtncRNAdb: plant transfer RNA-derived non-coding RNAs (tncRNAs) database pubmed.ncbi.nlm.nih.gov/35462956/ BRIC-NIPGR Department of Biotechnology