
Sam Kovaka
@samkovaka
Computational biology postdoc at JHU
ID: 348866045
05-08-2011 03:56:33
20 Tweet
269 Followers
140 Following

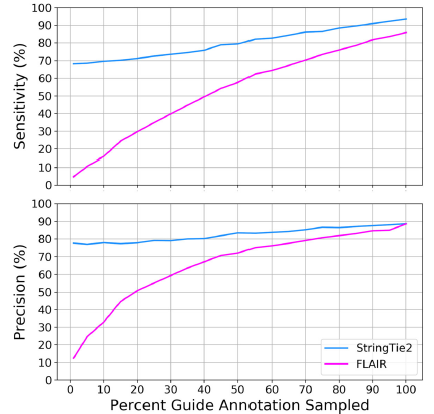
StringTie2 from Sam Kovaka, Mihaela Pertea and co. An update of the popular reference-guided transcriptome assembler, now includes the ability to assemble long reads. It's more accurate than other assemblers, while being quicker with lower memory requirements genomebiology.biomedcentral.com/articles/10.11…



Want to find out about how a class project turned into an Nature Biotechnology paper? Read this "Behind the Paper" blog post I wrote about the UNCALLED paper! bioengineeringcommunity.nature.com/posts/software…

A very simple yet very effective approach: High resolution copy number inference in cancer using short-molecule Oxford Nanopore sequencing. Great work with Timour Baslan Sam Kovaka Fritz Sedlazeck Sara Goodwin et al. biorxiv.org/content/10.110…



Need lots and lots of reads from your Oxford Nanopore for copy number analysis of cancer genomes? Try short molecule sequencing! Impactful project with Timour Baslan Sam Kovaka Fritz Sedlazeck Sara Goodwin et al. academic.oup.com/nar/advance-ar…


Thank you! So happy to finally (pretty much) be a doctor. Also excited to announce that I'll be staying at Hopkins as a postdoc with Michael Schatz!

Bioinformatics advancements unleash the full potential of long-read sequencing in various areas of genomics. A Comment from Michael Schatz (Michael Schatz) and colleagues highlights this thriving domain of active methods development. Johns Hopkins University nature.com/articles/s4159…


Very excited to announce "Uncalled4 improves nanopore DNA and RNA modification detection via fast and accurate signal alignment" by Sam Kovaka et al. biorxiv.org/content/10.110…

