
Sabina Leanti La Rosa
@sabinallarosa
Associate Professor of Microbiology @UniNMBU. Fond of carb degradation by gut #microbiomes, #multiomics, #CAZymes,#prebiotics. Editorial Board Member @CommsBio
ID: 1007297608413581313
https://www.nmbu.no/ans/sabina.leantilarosa 14-06-2018 16:23:59
1,1K Tweet
1,1K Followers
826 Following

The overlooked biodiversity loss! In a team of bright minds (Naia Morueta-Holme, Caroline Winther-Have, JacobAgerbo 🐟 🦠 & Tom Gilbert), we talk about the importance and consequences of also considering the invisible components of what we are losing. Center for Evolutionary Hologenomics Holobiont/Hologenome authors.elsevier.com/a/1jgYOcZ3X052K

Check out our new perspective in The ISME Journal, Diversity alone does not reliably indicate the healthiness of an animal microbiome. Claire Williams Toby Hammer [1/7] doi.org/10.1093/ismejo…




Join my group in beautiful Copenhagen NNF Center for Basic Metabolic Research (CBMR). A 3-year international postdoc position is available in my group. Topic is developing multi-omics foundation models bit.ly/3T98er5 #jobalert #postdoc #bioinformatics #deeplearning #multimodal #integration


Very excited to share our latest preprint! Led by a great student Charlotte A. Clayton, we show how bowel prep increases vulnerability to Salmonella as well as IBD pathobiont infection. We delve into mechanisms and the many facets of this common gut perturbation. shorturl.at/lOoe4

Happy to share our new paper on the characterization of MSTV1, an archaeal virus infecting Methanobrevibacter smithii, the dominant archaeon in the human gut. This was a beautiful collaboration between Mart Krupovic and Simonetta Gribaldo labs. 👇🧵nature.com/articles/s4146…

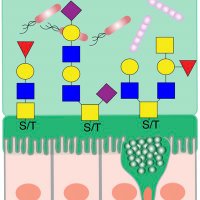
🚨New Paper Alert! Over 8 years in the making (including 2 in peer review 😬) - I could not be happier to share our genome-wide association study of #HumanMilk Oligosaccharides in the CHILD Cohort Study - published today in Nature Communications. 🧬🤱🏻🫁 #Tweetorial 🧵 nature.com/articles/s4146…




ICYMI, out now in nature: Global marine microbial diversity and its potential in bioprospecting Thomas Mock UEA School of Environmental Sciences UEA Research #microbiome #biodiversity nature.com/articles/s4158…

Interested in the absolute quantification of bacterial strains in fecal samples? My group has used qPCR for more than 10 years with good success, and we now published a step-by-step protocol for the design of strain-specific qPCR assays in Microbiome. doi.org/10.1186/s40168…


A convenient nail for the SingleM hammer. See Raphael Eisenhofer's 🧵x.com/R_Eisenhofer/s… Thanks also Antton Alberdi Centre for Microbiome Research and efficient reviewers/editor.

📢Our new paper is out! We show that gene regulation in breastmilk is controlled by maternal genetics, and impacts the composition of the infant gut microbiome cell.com/cell-genomics/… Work by Kelsey Johnson, in collab w/Frank Albert Ellen Demerath and many others







