Hamim Zafar
@quasarzafar
Assistant Professor and Early Career Fellow, DBT/Wellcome Trust India Alliance, IIT Kanpur, India
IYBA Awardee
Former Lane Fellow, Carnegie Mellon University
ID: 120360258
https://hamimzafar.wixsite.com/home 06-03-2010 06:12:05
2,2K Tweet
537 Followers
245 Following

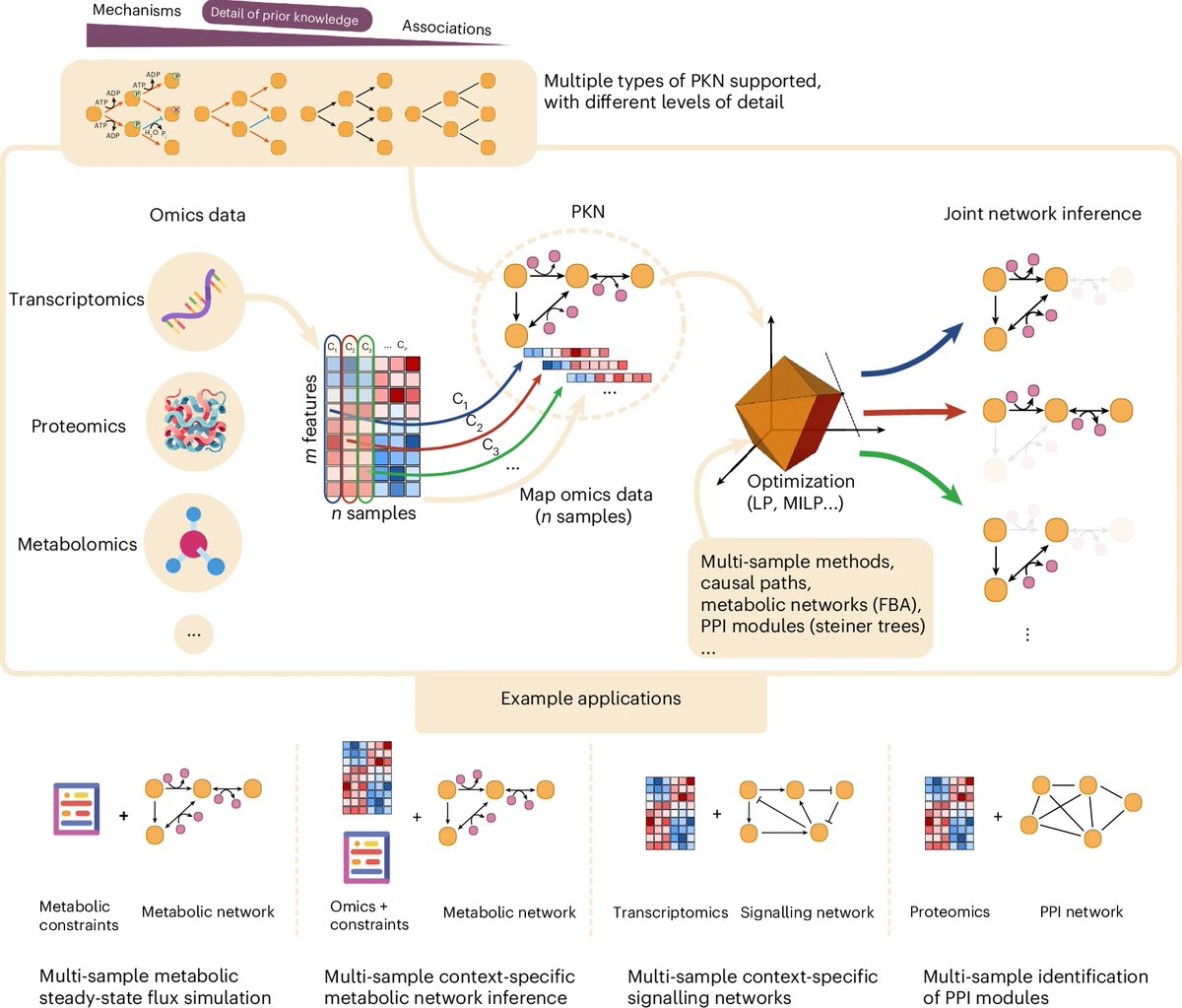
...but we're not there yet and at Arc Institute we are putting together all the pieces to make this happen! Check out: State: arcinstitute.org/manuscripts/St… cell-eval: github.com/ArcInstitute/c… scBaseCount: biorxiv.org/content/10.110…


The FUTURE is NOW! 🚨 Whole-Transcriptome Spatial Genomics with OUT-OF-THE-BOX analysis starring Helena Lucia Crowell. 🔬 Spatial genomics has now reached FULL-transcriptome resolution, enabling spatially resolved single-cell analysis at million cell scale. With this, we’re rethinking how


Very excited to share our preprint on #AI directed Deep Visual Proteomics (DVP) on #PancreaticCancer precursor lesions (PanINs), in collaboration with OmicVision Andreas Mund Thousands of proteins from as few as ~100 precisely microdissected cells in archival samples!






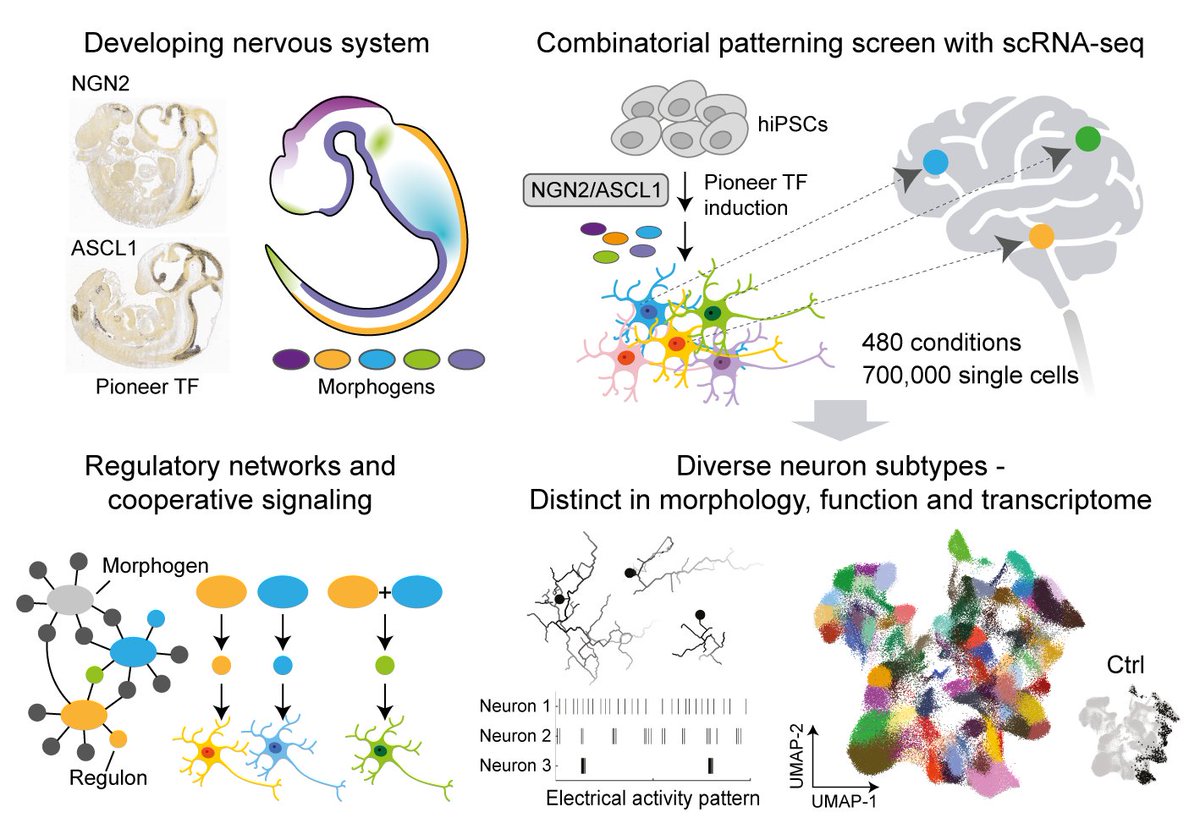
Neuron programming! Pro-neural TFs + 480 morphogen conditions + scRNA-seq --> Diverse iN subtypes of forebrain, midbrain, hindbrain, spinal cord, and PNS. science.org/doi/10.1126/sc… Hsiu-Chuan Lin Jasper Janssens Treutlein lab Science Magazine D-BSSE, ETH Zurich #NGN2 #ASCL1

1/11 Our study on subclonal immune evasion in non-small cell lung cancer has just been published in Cancer Cell cell.com/cancer-cell/fu…


I’m happy to share our latest work on tumour necrosis! During my time in the Mikala Egeblad lab, we found that tumour necrosis is not a passive phenomenon secondary to tumour growth, but an active phenomenon driven by neutrophils and NETs! Thread below: (1/13) nature.com/articles/s4158…

This work was led by stellar postdoc Aditya Pratapa, co-supervised by @tatalab-duke.bsky.social , and with fantastic collaborators @kevinmbyrd and Rajkumar Savai! Here's the preprint: biorxiv.org/content/10.110… The code will be here: github.com/rohitsinghlab/…