
Picotti Lab
@picotti_lab
Mass spectrometry-based #proteomics lab at @IMSB_ETH studying protein conformational changes & their effect on cell physiology. Tweets are from lab members
ID: 1187702683869859840
https://imsb.ethz.ch/research/picotti.html 25-10-2019 12:10:22
671 Tweet
3,3K Followers
434 Following

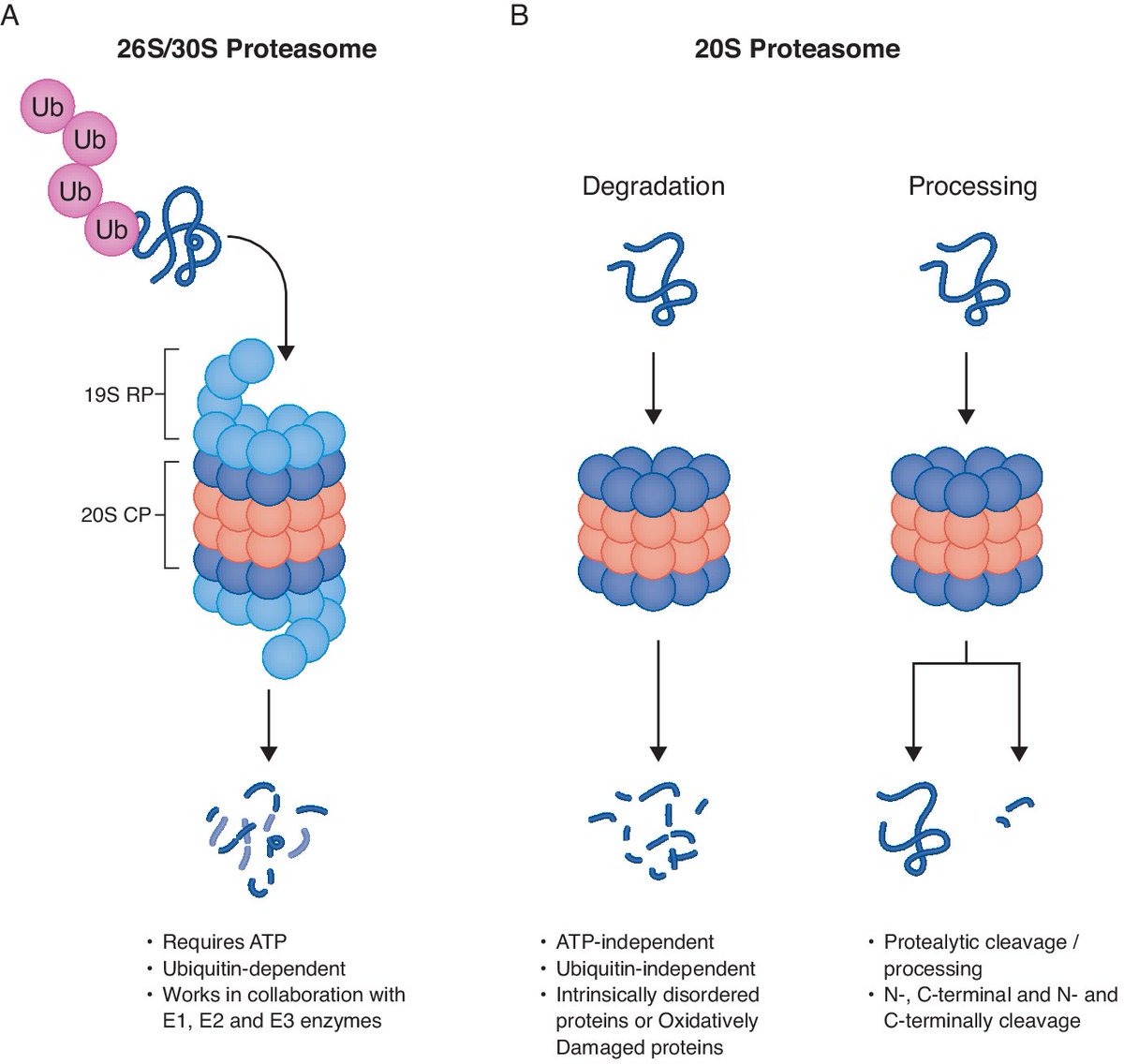
A systematic investigation of 20S #proteasome substrates reveals 100s of substrates & differential functionality of naïve and oxidized 20S proteasome complexes ➡️ embopress.org/doi/full/10.10… Picotti Lab IMSB_ETH Michal Sharon Weizmann Institute #proteomics #massspectrometry
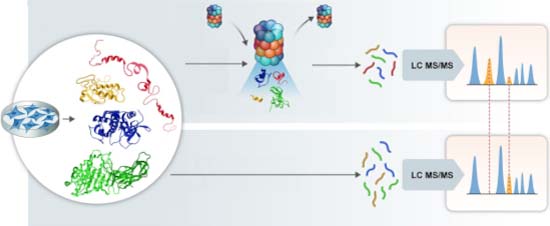

Our paper with Michal Sharon is now live Mol Syst Biol. We identified hundreds of substrates of the 20S proteasome, enriched in RNA- and DNA-binding proteins with IDRs. A variety of substrates showed site-specific cleavage, suggesting functional products. embopress.org/doi/full/10.10…


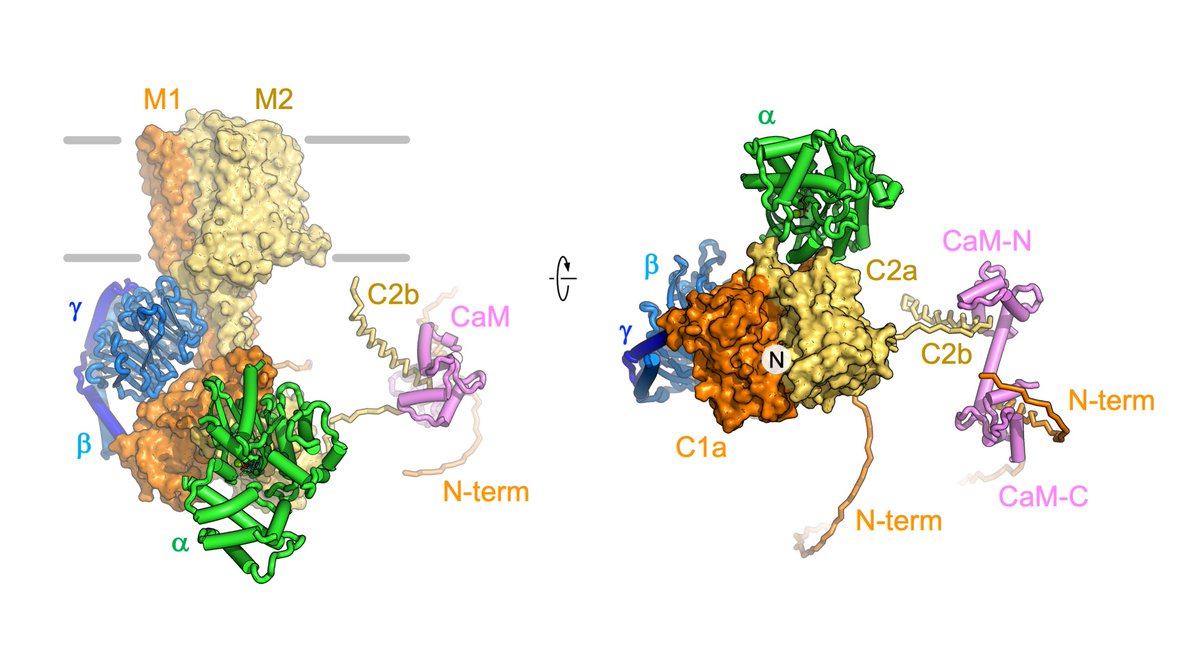
The paper by Korkhov Lab, together with Picotti Lab, Alexander Leitner and Federico Uliana, now out in EMBO Reports, describes the cryo-EM- and structural proteomics-based analysis of adenylyl cyclase AC8 and its macromolecular complexes. More: biol.ethz.ch/en/news-and-ev…

Publication by the Allain & Leitner Labs (IMSB_ETH, ETH Zurich) with 1st authors Tebbe de Vries & Mihajlo Novakovic (@tflut1,Mihajlo Novakovic) "Specific #protein-#RNA interactions are mostly preserved in biomolecular #condensates" Science Advances bit.ly/4cfo0sb #MS #LLPS

Great talk by Tetiana Serdiuk at the AD/PD conference on biochemical differences between amyloid strains linked to different alpha-synucleinopathies. AD/PD - Advances in Science & Therapy



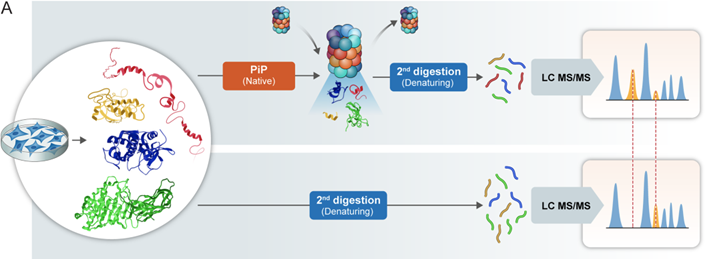
N&V by Seth S. Margolis on the study by Picotti Lab & Michal Sharon developing an advanced #proteomics method (PiP-MS) to comprehensively explore 20S #proteasome substrates in the human proteome ➡️ embopress.org/doi/full/10.10…


High resolution protein complex analysis beyond AP-MS by the Gstaiger group: nature.com/articles/s4159…. Nice collaborative adventure at IMSB_ETH from the @MGstaigerlab members @FabianFrommelt , Andrea Fossati , Federico Uliana and from Bernd Wollscheid lab members



Our paper describing a new method (FLiP-MS) for the global profiling of protein complex dynamics is out Nature Biotechnology ! Read here👉: doi.org/10.1038/s41587… 🔬 Highlights: - Uses an experimental library of peptide markers integrated with a structural proteomics workflow. -
