
Denys Oliinyk
@oliinyk_denys
Postdoc at @labs_mann | @fmeierX & @lindaunobel alumni
ID: 1249727590308470784
13-04-2020 15:54:19
78 Tweet
113 Takipçi
215 Takip Edilen



Do you want to push the boundaries of mass spectrometry-based phosphoproteomics and make a difference in virology? We have a 3-years PhD position available funded by DFG public | @[email protected] in collaboration with Systems Arbovirology. Please RT and reach out if interested.

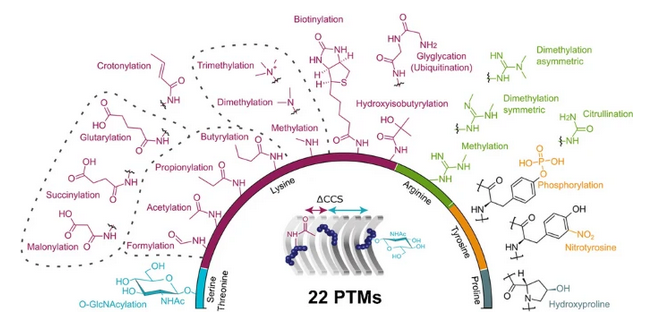
Peptide collision cross sections of 22 post-translational modifications by Florian Meier et al Florian Meier Denys Oliinyk Uniklinik Jena Bleiholder Group FSU Chemistry & Biochemistry Florida State University #peptides #PTMs #OpenAccess link.springer.com/article/10.100…

Excited to see this exciting and thourough story, led by @A_Will_tweets, out in Anal Bioanal Chem! Check it out!


And the big THANK YOU goes this time to Florian Meier and colleagues for this great cover! 🎉 @A_Will_tweets Florian Meier Denys Oliinyk Uniklinik Jena Bleiholder Group FSU Chemistry & Biochemistry Florida State University Read here their Paper in Forefront: link.springer.com/article/10.100…




What a great honor to receive the Am Soc for Mass Spec Graduate Student Award! A big thank you to Florian Meier and the whole group for the immense support, can’t wait to celebrate other recipients in June!

Thrilled to finally see µPhos published in Mol Syst Biol! A big thank you to Florian Meier, Sean Humphrey and all other coauthors for this exciting journey! Check out our greatly improved and extended story on how to unlock the true power of phosphoproteomics: embopress.org/doi/full/10.10…

Spectronaut solves many of the challenges of large-scale proteomics studies. #Spectronaut user Florian Meier (Uniklinik Jena) shares with us why Spectronaut is his software of choice for PTM studies at scale. Get in touch for a free trial: ow.ly/eI2O50SuCfp


Introducing @PulsPhos: our new method to quantify phosphorylation lifetimes in living cells. See how it reveals key protein dynamics! #Phosphoproteomics #CellBiology Yansheng Liu Pedro Beltrao Julian van Gerwen @[email protected] Nisha Hemandhar Kumar biorxiv.org/content/10.110…





AATD affects thousands, causing liver & lung disease via protein aggregation. Our new preprint used #DeepVisualProteomics to study liver cells and reveals key AATD insights with clinical implications . Video and tweetorial Florian Rosenberger #Alpha1Awareness bit.ly/4hzl64o




