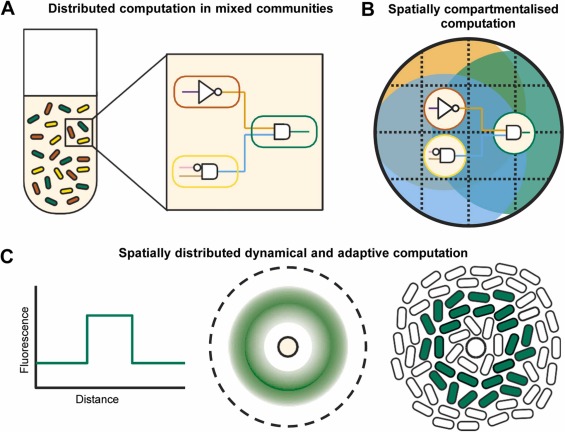
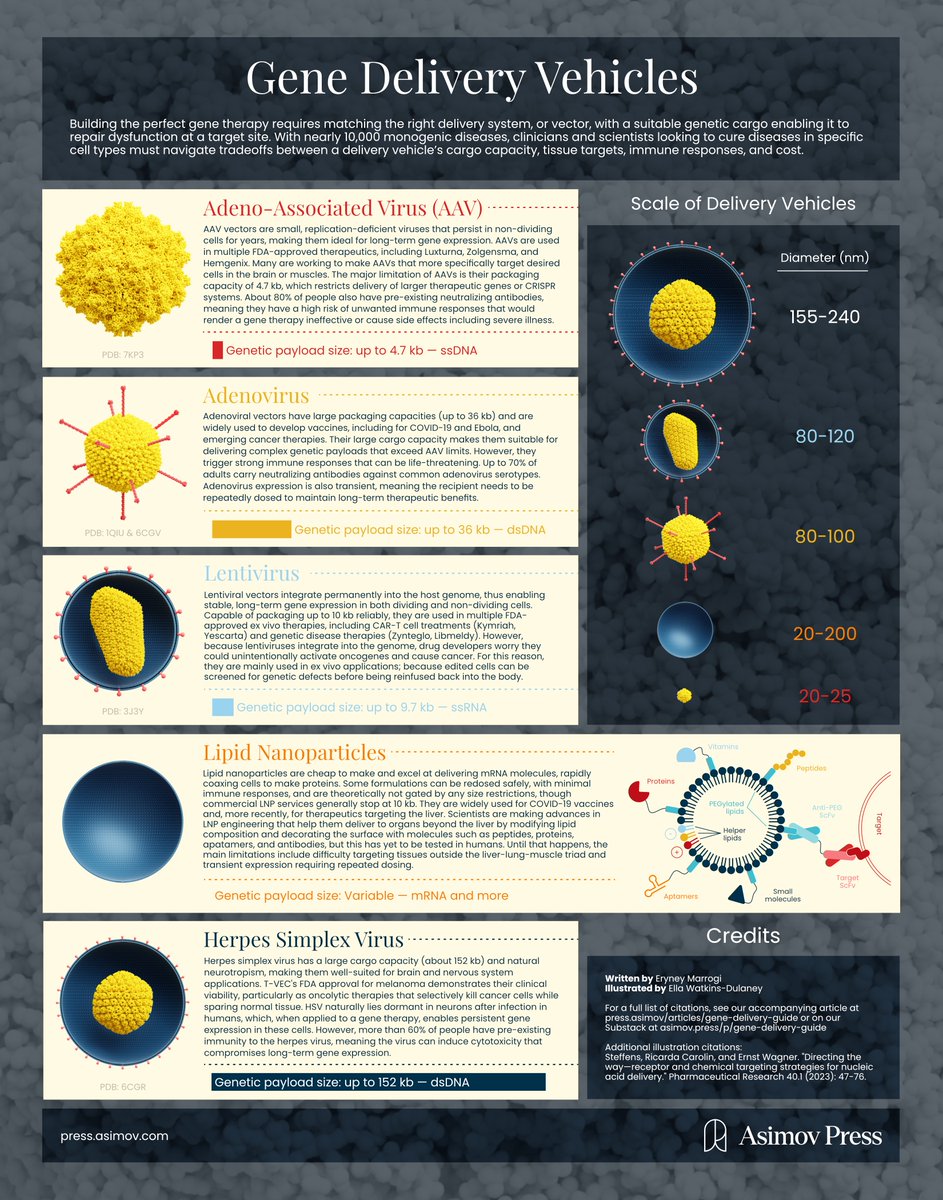
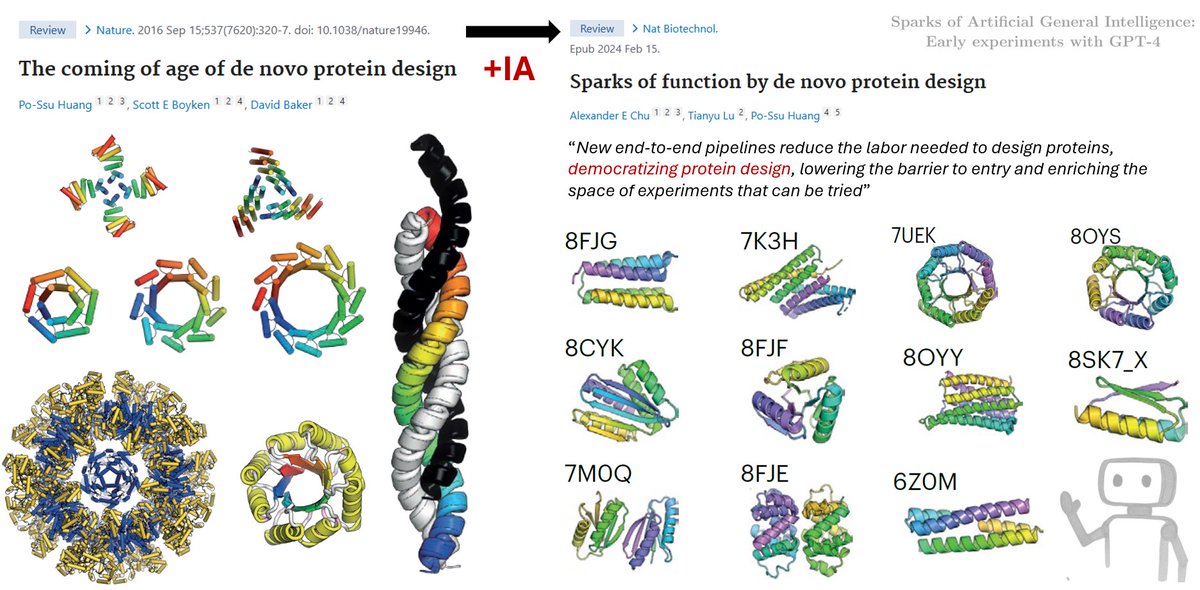
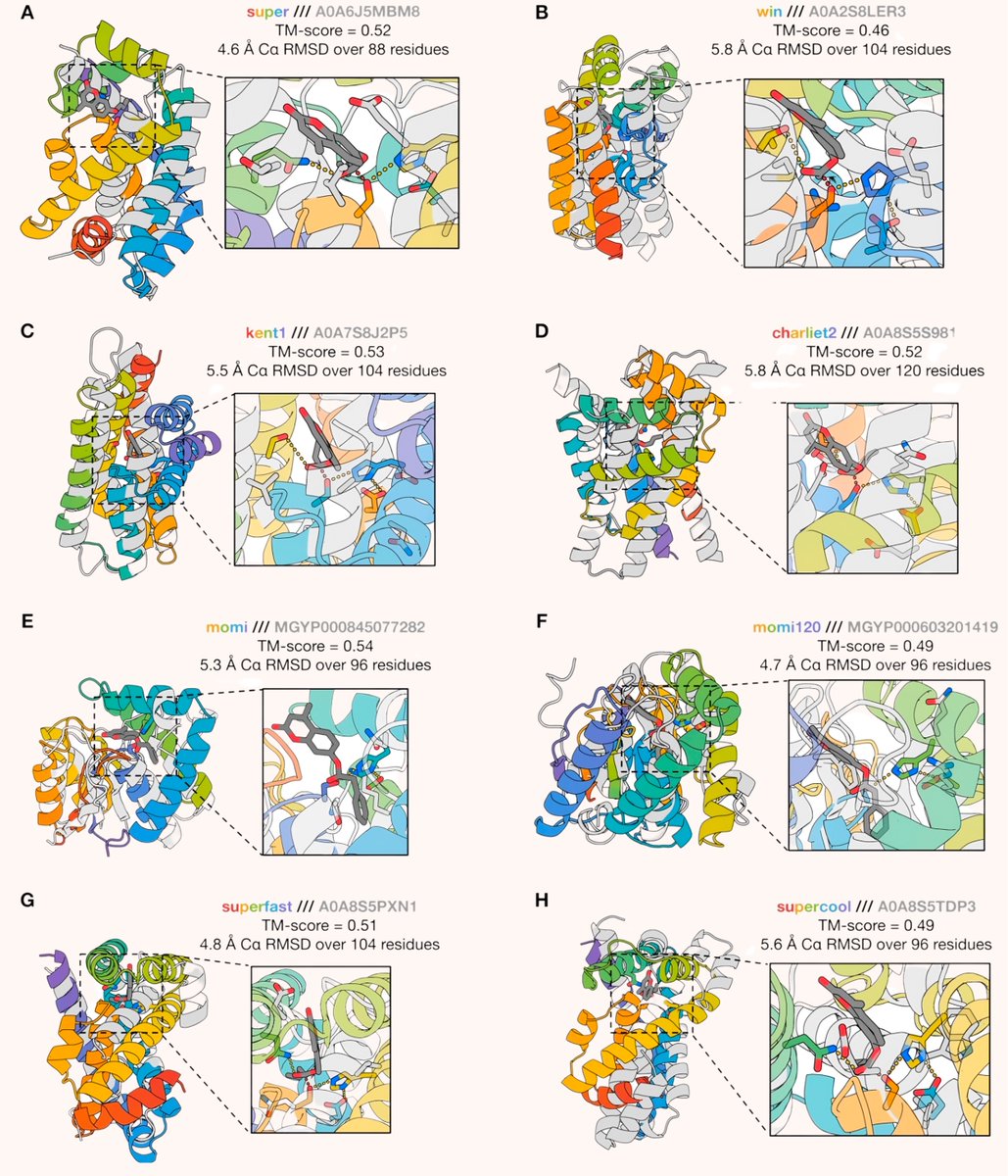
Noot
@nootlabs_
🧬 SynBio PhD Candidate
🏠 Setting up my home lab
🧫 Follow me for updates and tips on low-cost lab equipment and protocols
#diybio #opensource #labonthecheap
ID: 1590006376205496320
https://nootlabs.substack.com/ 08-11-2022 15:40:56
625 Tweet
688 Takipçi
160 Takip Edilen









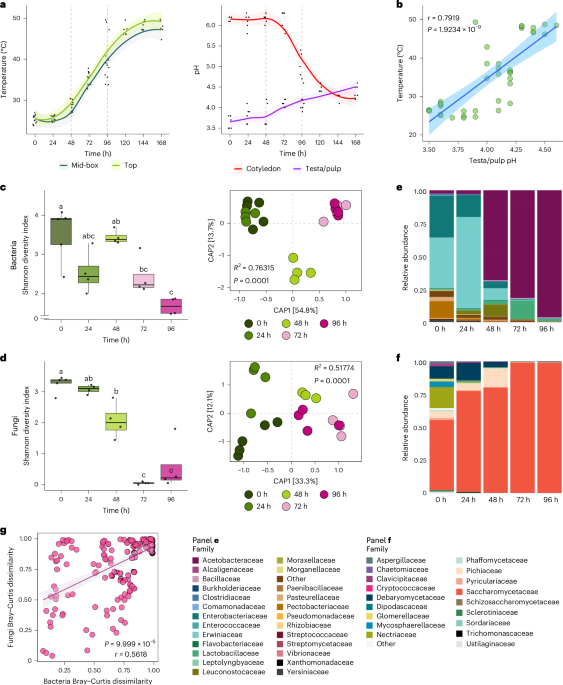
New paper from our lab on synthetic genome work in yeast is out - Iterative SCRaMbLE for Engineering Synthetic Genome Modules and Chromosomes. Exciting project led by Jane (Xinyu) Lu in our group, now online Nature Communications nature.com/articles/s4146…