
Nathan Abell
@n_s_abell
computational genetics @OctantBio | previously @StanfordMed and @UTAustin | 🏳️🌈
ID: 936683790348333056
http://nsabell.github.io 01-12-2017 19:49:53
100 Tweet
178 Followers
200 Following


1/ Learning compound-protein interactions (CPI) w/ sequence & compound alone is tantalizing, but time and time again, CPI models fail to beat simple baselines. Does this paper do so successfully? An analysis from Sam Goldman :



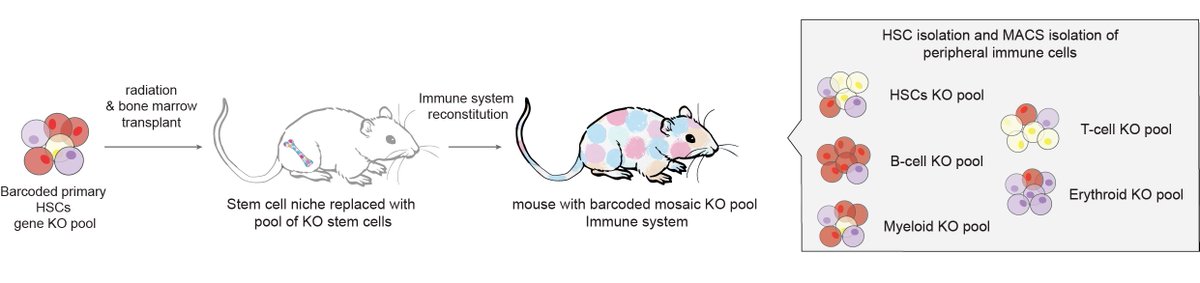
Excited to share our work on in vivo hematopoietic stem cell CRISPR screens to study hematopoiesis and the aging immune system in the Tony Wyss-Coray lab w/ Wilkinson Group at Oxford & Nakauchi lab at Stanford! biorxiv.org/content/10.110… Find our screen data at hematopoiesiscrisprscreens.com

Hiring a scientist into our Deep Mutational Scanning team for an exciting project in partnership with Robert Plenge and BMS. If you are into reporter & cell engineering, multiplexed screens & how to use DMS for identifying and drugging targets, please apply! octant.bio/jobs?gh_jid=47…

You'd be working with some great scientists including Diane Dickel who leads the team, and Conor Howard, Erin Thompson Nathan Abell et al. If you have any questions, please feel free to reach out to any of us.

Excited to share my PhD work in the @TobiasMeyerLab, out now in Molecular Cell! “CDT1 inhibits CMG helicase in early S phase to separate origin licensing from DNA synthesis” 1/11 cell.com/molecular-cell…

We held a reading group on Transformers (watched videos / read blog posts / studied papers by Lucas Beyer (bl16) Andrej Karpathy Chris Olah Aman Arora Jay Alammar Sasha Rush et al.), and now I _finally_ roughly understand what attention does. Here is my take on it. A summary thread. 1/n










Excited to open up work on improving the power of Deep Mutational Scanning and applying it to drug discovery. I’ll comment on the preprint itself, which is work from a few years ago on DMS of MC4R, as well as talk about how this has evolved Octant Bio over time. 1/🧵





![Sasha Gusev (@sashagusevposts) on Twitter photo Another outlier: In [Okbay et al. 2022] the relative accuracy of an Education PGI in (UK Biobank) individuals with African ancestry was ~0%, far lower than expected from genetic differences. This is the finding that makes me wonder if *any* EA GWAS hits are direct effects at all. Another outlier: In [Okbay et al. 2022] the relative accuracy of an Education PGI in (UK Biobank) individuals with African ancestry was ~0%, far lower than expected from genetic differences. This is the finding that makes me wonder if *any* EA GWAS hits are direct effects at all.](https://pbs.twimg.com/media/FvijRfzX0AIl94M.jpg)

