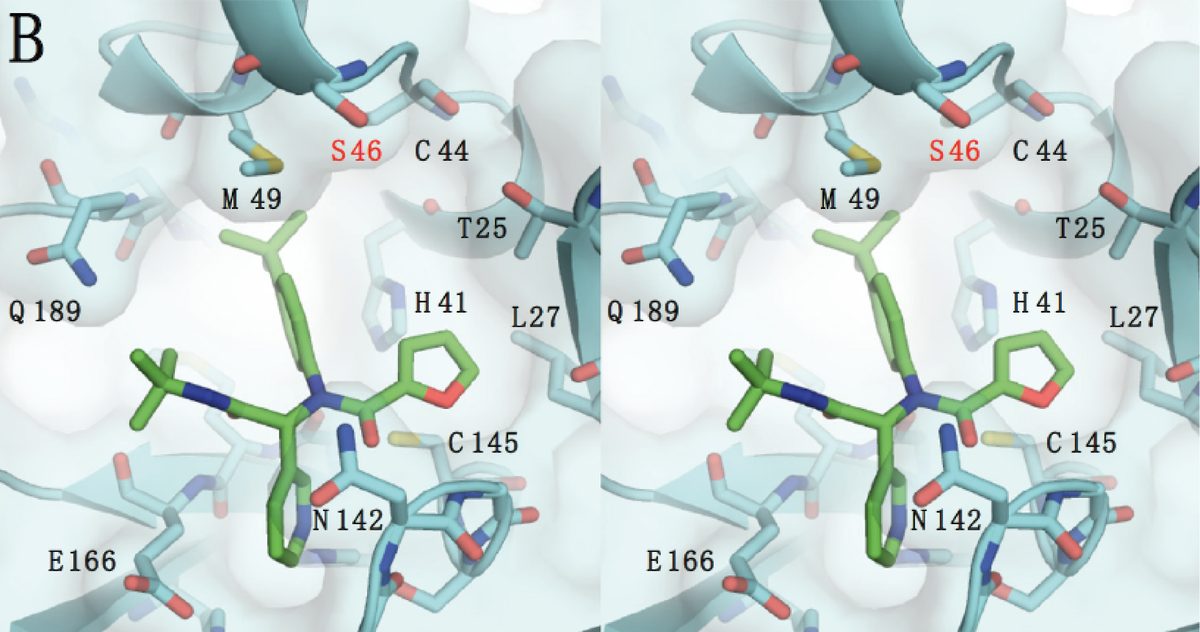
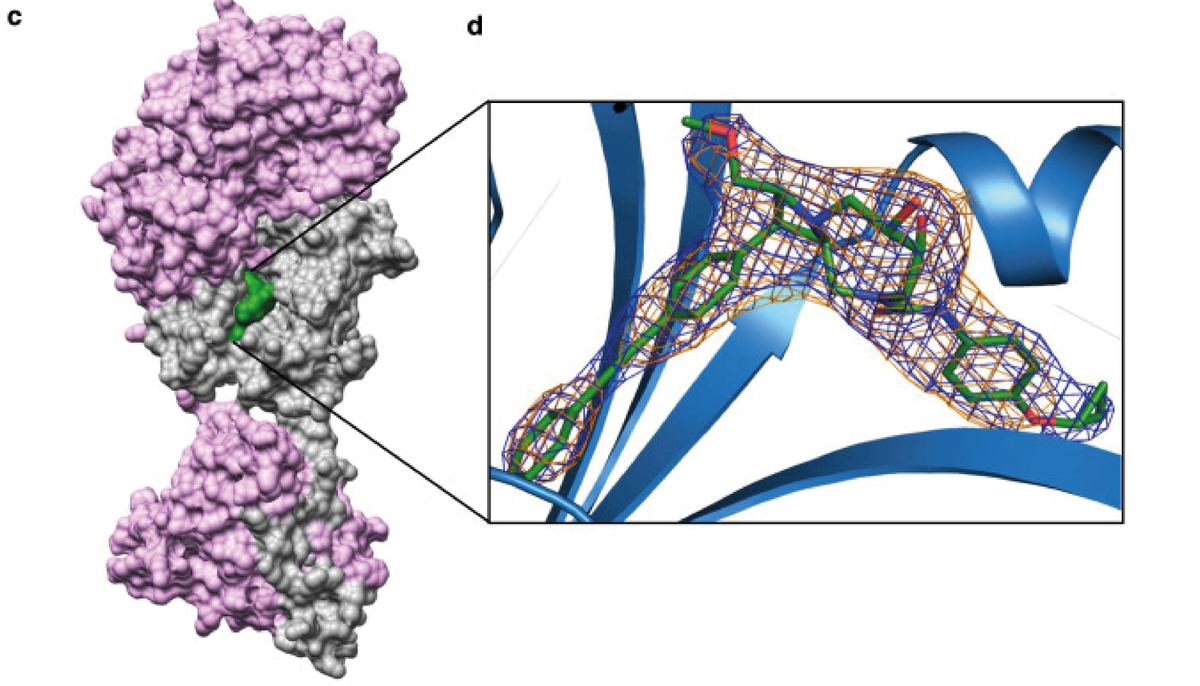
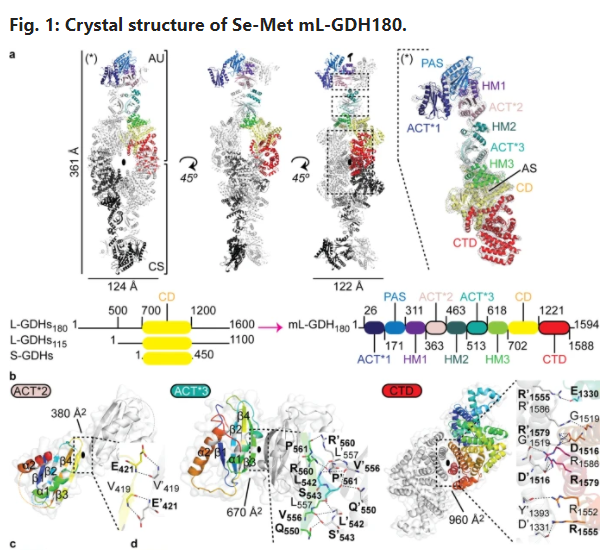
Molecular Dimensions
@moleculard
The official Twitter account of Molecular Dimensions
ID: 516393278
http://moleculardimensions.com 06-03-2012 10:03:12
1,1K Tweet
1,1K Followers
680 Following







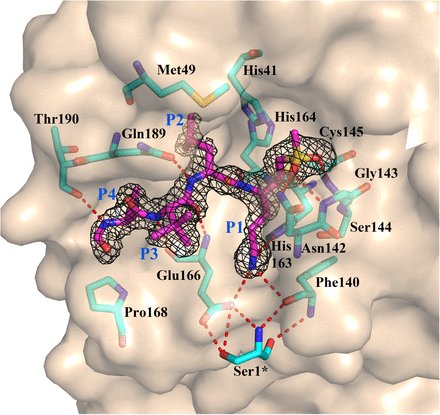
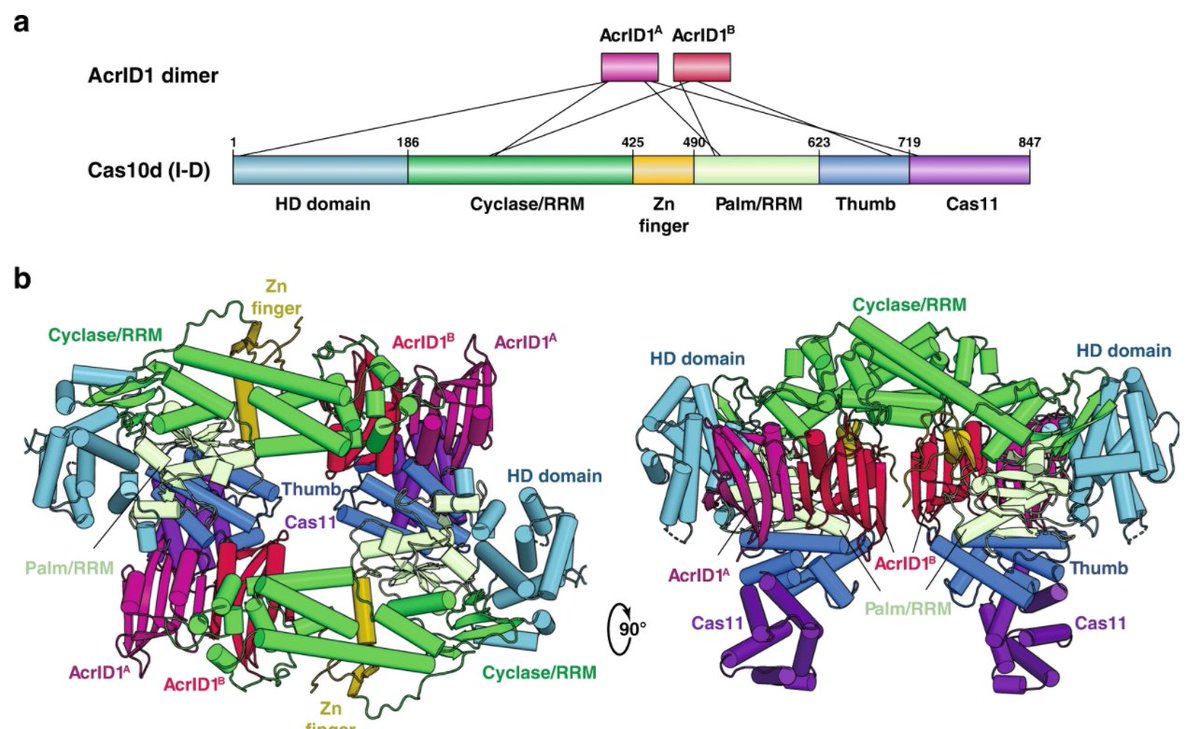
Rodnin et. al. from KU Medical Center examined the structure of Diptheria Toxin with crystal obtained using Molecular Dimensions drop crystallisation plates doi: 10.3390/toxins12110704 #Calibre Scientific Find all our plates here: hubs.ly/H0DjWqk0



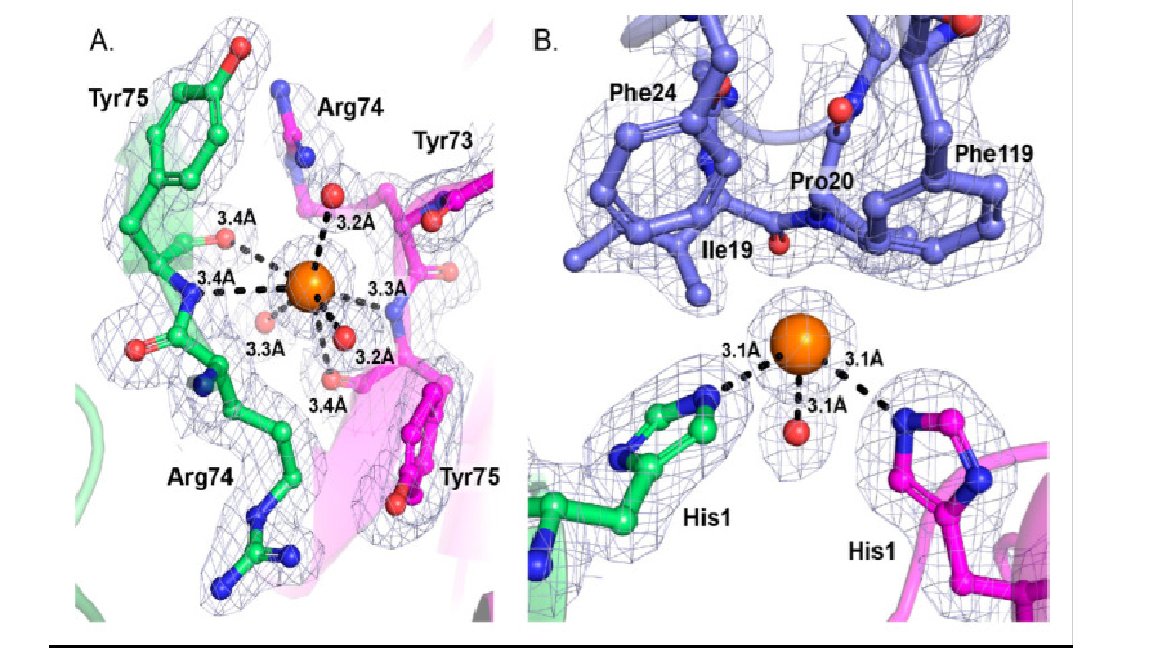
Researchers at Sheffield crystallised the A component (SmhA) of a tripartite pore-forming toxin using Molecular Dimensions Screens: JCSG+, Pact premier, ProPlex and Morpheus doi: 10.1107/S2053230X20013862 #CalibreScientific Find out about all our screens here: hubs.ly/H0DjZ770


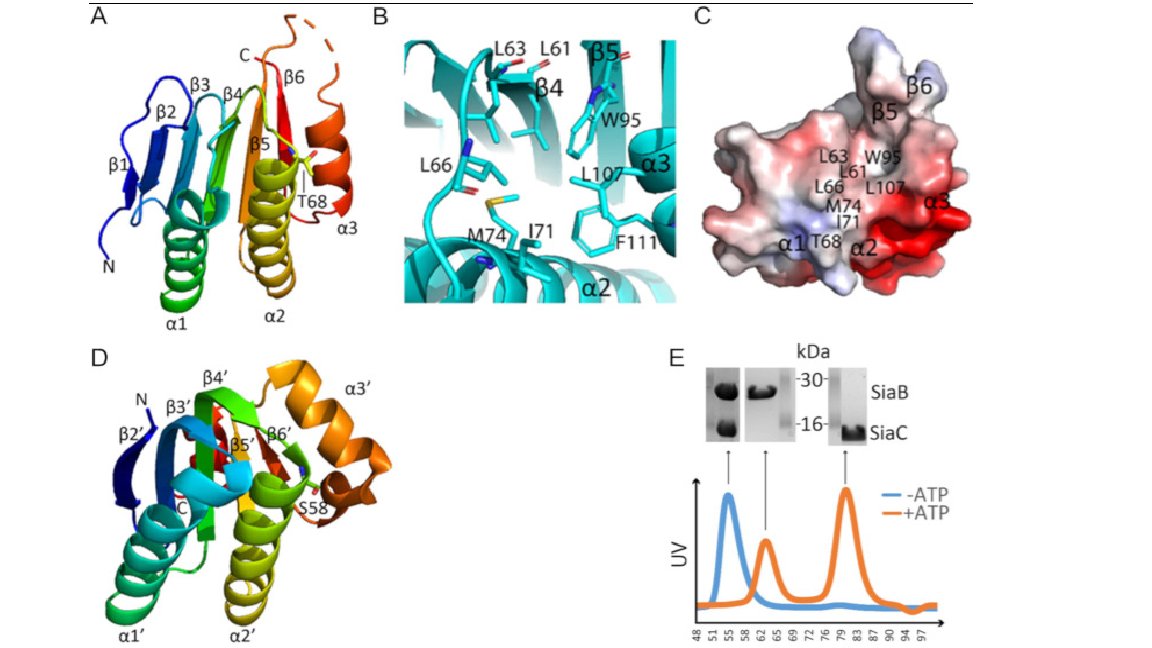
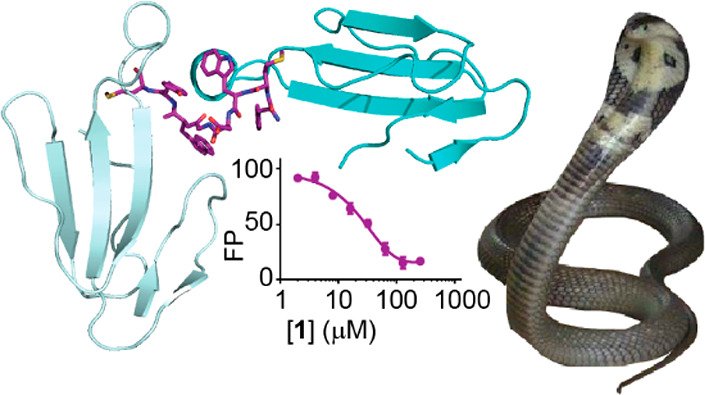
Crystals of the SiaABC domain grown using the Molecular Dimensions JCSG+ and Morpheus screens helps researchers to better understand biofilm growth doi: 10.1371/journal.pone.0241019 Find out more about our screens here: hubs.ly/H0DkpkH0