
Ming Wang
@ming0325
PI @Institute of Zoology, CAS
Interested in plant epigenetics, epigenome editing, and plant pathogen interactions
ID: 207790428
http://english.ioz.cas.cn/sourcedb/scs/202310/t20231030_431170.html 26-10-2010 01:26:23
17 Tweet
28 Takipçi
93 Takip Edilen

My latest story looks at how research from the 1960s has inspired scientists like UC Riverside's Hailing Jin to design tomatoes that release exRNA as a fungicide. She's now making crop sprays that contain silencing RNA. Have a read in this Nature Outlook: nature.com/articles/d4158…

CRISPR-CAS mediated transcriptional control and epi-mutagenesis Jason Gardiner Basudev Ghoshal Ming Wang Steve Jacobsen #PlantSci academic.oup.com/plphys/advance…
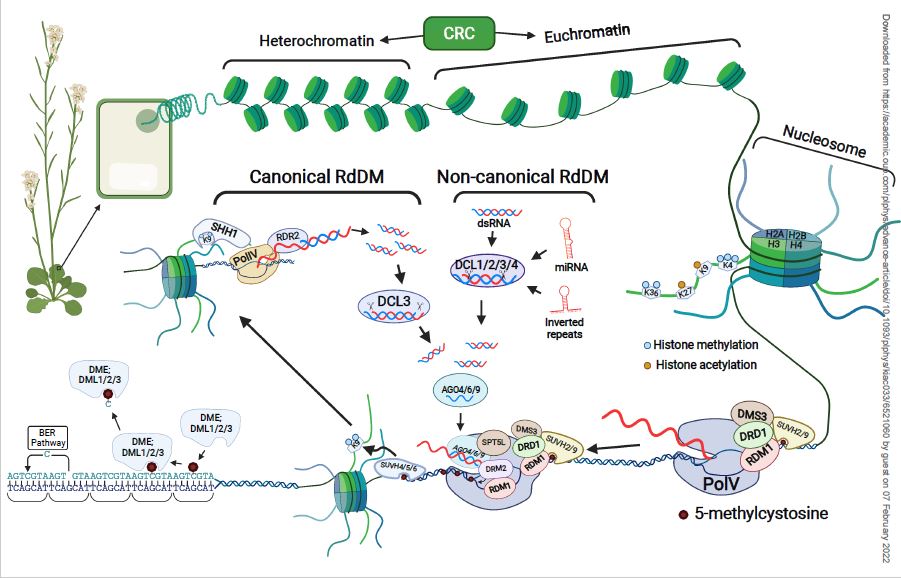








Cool epigenome editing screen from Steve Jacobsen. They found the first protein I ever published on: JMJ14 (K4 demethylase). JMJ14 leads to silencing of the target, but no change in DNAme (or K27me3). We saw in the jmj14 mutant there was a DNAme defect 🤔 nature.com/articles/s4147…



Excited to share our work published in Nature Plants today rdcu.be/d9rPo Histone H3 lysine 4 methylation recruits DNAdemethylases to enforce gene expression inArabidopsis. Thank you Steve Steve Jacobsen, Yan and all the co-authors for the support! #Epigenetics


Congratulations, mary droser and Hailing Jin of UC Riverside on being elected National Academy of Sciences members! nasonline.org/news/2025-nas-… UC Riverside CNAS






