Matthias Blum
@matthiasblum
@InterProDB/@PfamDB Development Project Leader at @emblebi. Interested in automation, gene regulation, data visualisation, and reinventing the wheel.
ID: 481972235
03-02-2012 11:32:25
326 Tweet
142 Takipçi
626 Takip Edilen


James Taylor died yesterday. Our dear James Taylor -> Galaxy began with you. We will make sure it continues. We have no words.



🥳 A new InterPro paper has been published in Nucleic Acids Res ! Find out about the new features and data update we have been working on in the past 2 years: academic.oup.com/nar/advance-ar…



We are delighted to announce the release of structural models for 6,370 families of unknown structure provided by Baker group Institute for Protein Design: xfam.wordpress.com/2021/03/03/fol… (funded by Biotechnology and Biological Sciences Research) #structureprediction #trRosetta #structuralbiology #deeplearning #AI







This illustration by Inna-Marie, inspired by Waddington’s second most famous drawing, shows with remarkable directness how MultiFate’s 3 transcription factors, interacting in combinations, produce a multistable epigenetic landscape.







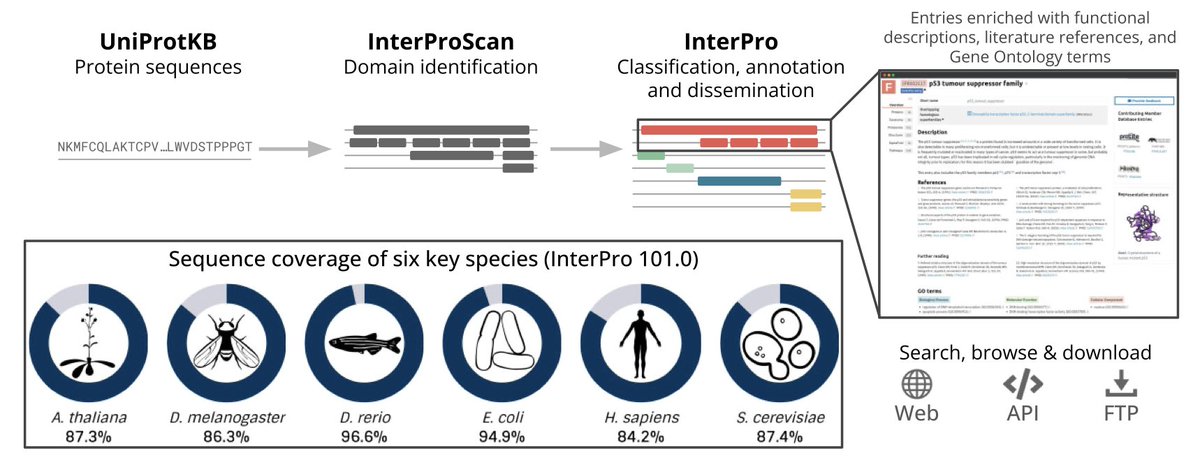
InterPro: the protein sequence classification resource in 2025 Nucleic Acids Res 1. InterPro version 101.0 provides a comprehensive resource for protein sequence classification, integrating over 84,000 predictive models (signatures) from 13 databases, covering more than 80% of