Leo Zang
@leotz03
Protein Designer | Share Reading Notes (AI+Protein/RNA/DNA) | @harvardmed @DeboraMarksLab | Hosting @ml4proteins | Curr. @Dyno_Tx
ID: 1763647865774350336
http://www.leozang.com 01-03-2024 19:30:25
1,1K Tweet
3,3K Followers
418 Following

🔥 Introducing BindEnergyCraft (BECraft), the BindCraft pipeline you know and love, now enhanced with an energy-based loss to boost in silico binder success! Thrilled to present this as an oral at ICML GenBio Workshop @ ICML25 next week 📄 Paper: arxiv.org/abs/2505.21241 🧵Thread⬇️




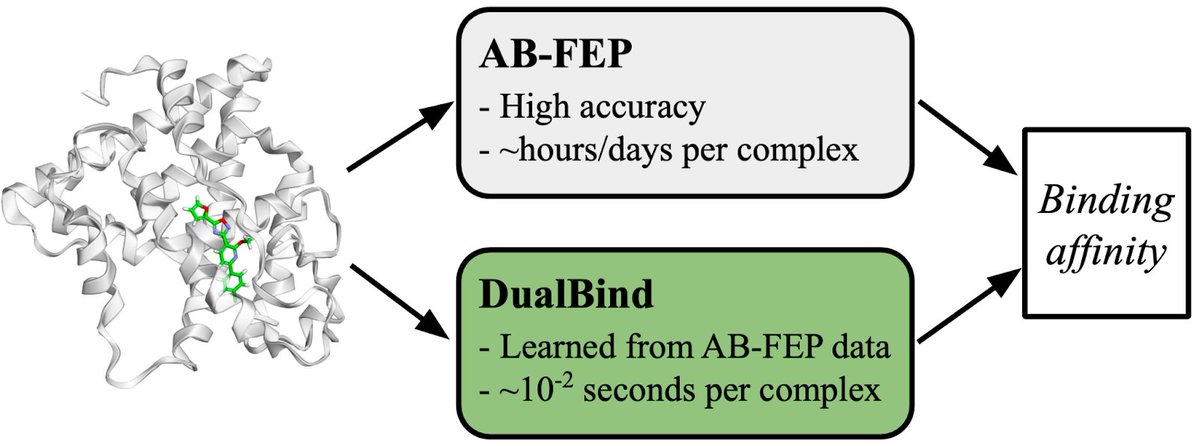
In collaboration with Schrödinger, we present DualBind trained on ToxBench 📚 8,770 ERα AB-FEP predicted complexes (RMSE ≈ 1 kcal mol⁻¹). At last, ML has the signal to learn true binding physics - DualBind already hits r = 0.84 without shortcuts. How could this reshape

Excited to share: “Learning Diffusion Models with Flexible Representation Guidance” With my amazing coauthors Cai Zhou, Sharut Gupta, Johnson Lin, Stefanie Jegelka, Stephen Bates, Tommi Jaakkola Paper: arxiv.org/pdf/2507.08980 Code: github.com/ChenyuWang-Mon…

Next Wed (7/23) at 4PM ET, Professor Possu Huang Possu Huang Lab will be giving a talk about his research! Sign up on our website for zoom links!








Next Wed (7/30) at 4PM ET, please join us for an exciting talk from Pranam Chatterjee about his research! Sign up on our website for zoom links!