Lukas Gerasimavičius
@l_geras
Research Fellow @jmarshlab | Institute of Genetics and Cancer, Edinburgh
ID: 1307626805055107073
20-09-2020 10:25:24
51 Tweet
84 Followers
186 Following

... which is another shameless self-promotion of our paper, recently published in EIC Jack Gilbert on the computational tools to dissect pombe metabolism: journals.asm.org/doi/10.1128/ms… 2/2

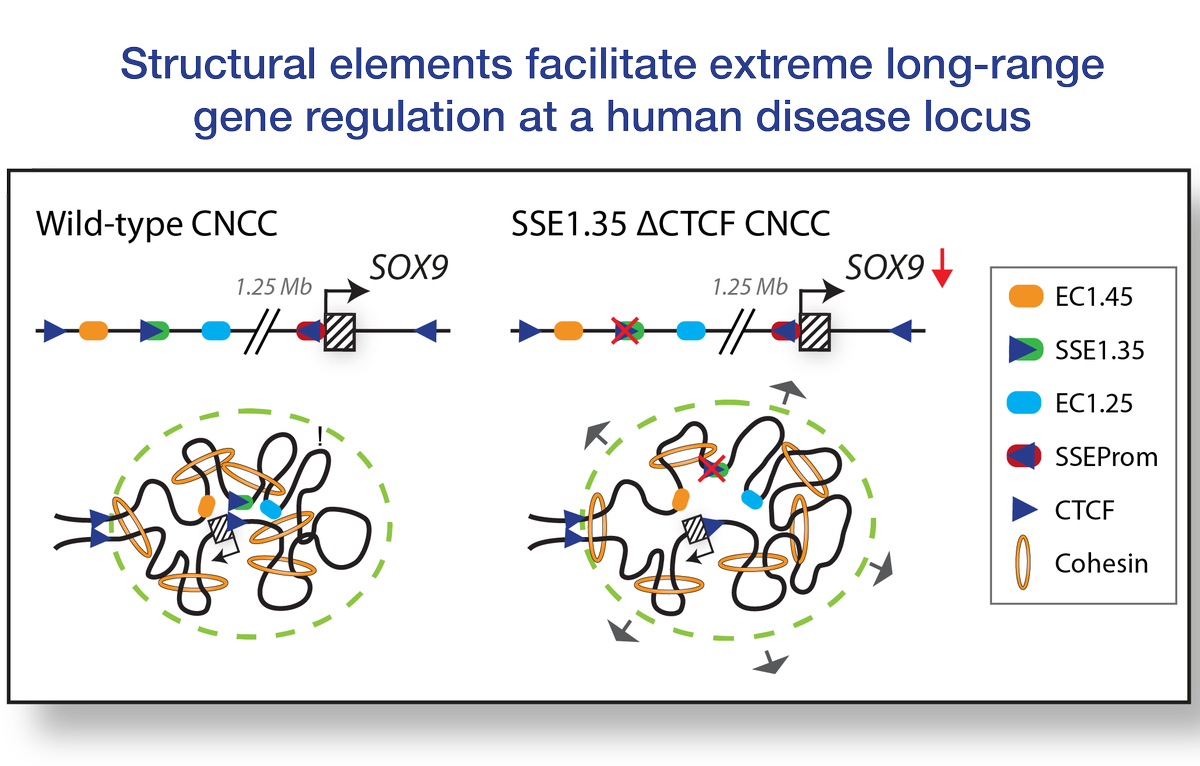
New #biorxiv preprint! We identify structural elements at a human disease locus that promote domain-wide interactions and facilitate extreme long range gene regulation. A fantastic collaboration with Liang-Fu Chen, Minhee Park, Tomek Swigut, @Aboettiger and Joanna Wysocka!

Excited to share this new preprint where we finally are able to address the longstanding hypothesis that cotranslational protein complex assembly can decrease the likelihood of dominant-negative mutations Mihaly Badonyi biorxiv.org/content/10.110…

Update to our 2020 study benchmarking variant effect predictors using deep mutational scanning data, featuring more VEPs and more DMS datasets. Ben Livesey biorxiv.org/content/10.110…

I am excited to share that our latest work on cotranslational protein complex assembly and its ability to counter the dominant-negative effect is now published in Science Advances. science.org/doi/10.1126/sc…


Excited to share our Perspective on the new AlphaMissense variant effect predictor tool from Google DeepMind Science Magazine science.org/doi/full/10.11…