Tobias Kroniger
@kronigert
Field Application Scientist @ Bruker Daltonics. Views are my own. he/him. #Proteomics #TeamMassSpec
ID: 1653820267024506882
03-05-2023 17:54:48
83 Tweet
68 Takipçi
149 Takip Edilen


📢Preprint alert Metaproteomics Initiative GutMicrobiota Health Host-Microbe Interactions Austrian Microbiome Mass Spec Pro Proteomics A quantum leap in microbiome research, metaproteomics, and proteomics biorxiv.org/cgi/content/sh… 🧵, below


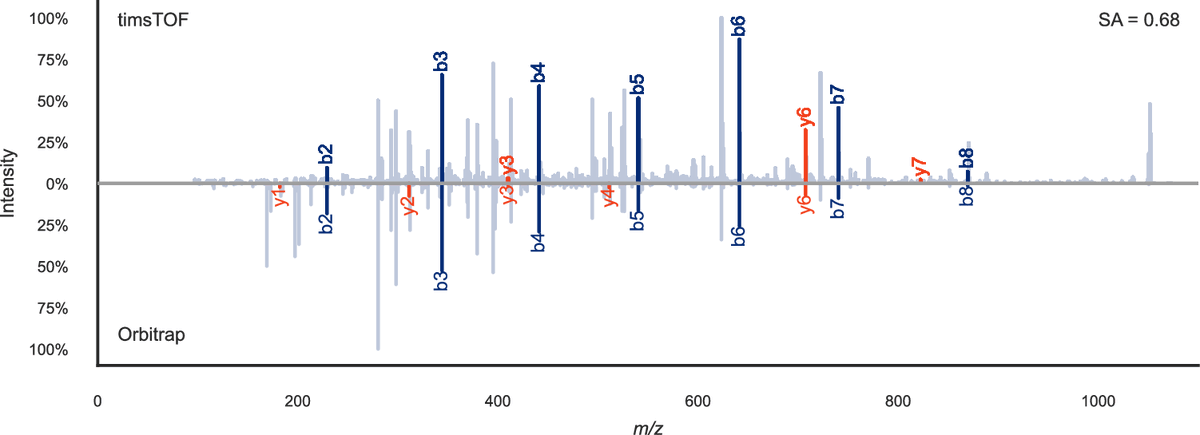
Technical Note and Tutorial for the analysis of XL-MS data with PASEF, MeroX, ProXL, and Skyline! pubs.acs.org/doi/10.1021/ac… Only possible thanks to the support from Andrea Sinz Lab and the patience and willingness from Michael Riffle and the Skyline Project dev team!










Thrilled to finally see µPhos published in Mol Syst Biol! A big thank you to Florian Meier, Sean Humphrey and all other coauthors for this exciting journey! Check out our greatly improved and extended story on how to unlock the true power of phosphoproteomics: embopress.org/doi/full/10.10…


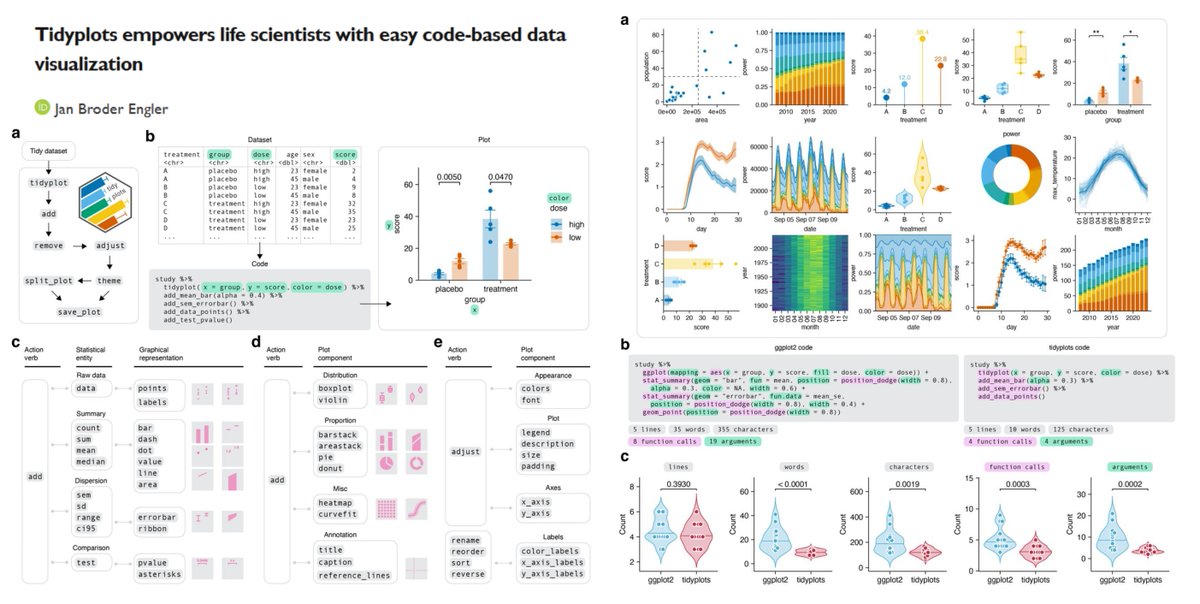
tidyplots Time to say goodbye to ggplot2?🫡 "a significant reduction of code complexity" vs ggplot2 cran.r-project.org/web/packages/t… Jan Broder Engler bioRxiv 2024 biorxiv.org/content/10.110…