
K-P Wu
@kpwu11
Structural biologist and biophysicist. Associate Research Fellow, Institute of Biological Chemistry, Academia Sinica, Taiwan.
ID: 1009404887090622464
http://kpwulab.com 20-06-2018 11:57:33
501 Tweet
205 Takipçi
193 Takip Edilen




Introducing Interactive Cell Biology, a complete course solution—no textbook needed. In partnership with Macmillan Learning, delivered through Achieve. Check it out today! Visit: macmillanlearning.com/college/us/pre… to learn more. #cell #biology #education #animation

I was happy to write a nature News and Views on the amazing discoveries by Brian Liau & Ning Zheng lab on how a molecular glue unexpectedly mimics the effect of cancer mutations. Such cool science and with implications to drug discovery. Nature News & Views nature.com/articles/d4158…


Structural biology is in an era of dynamics & assemblies but turning raw experimental data into atomic models at scale remains challenging. Minhuan Li and I present ROCKET🚀: an AlphaFold augmentation that integrates crystallographic and cryoEM/ET data with room for more! 1/14.



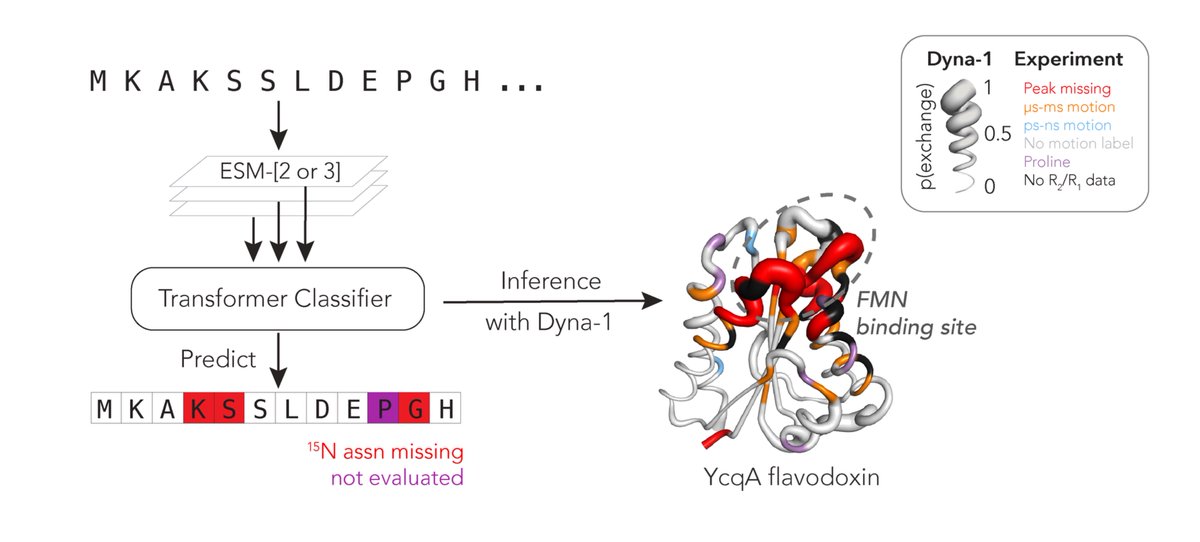
Protein dynamics was the first research to enchant me >10yrs ago, but I left in PhD bc I couldnt find big expt data to evaluate models. Today w Gina El Nesr, I'm thrilled to share the big dynamics data I've been dreaming of, and the mdl we trained: Dyna-1. rb.gy/de5axp


Protein function often depends on protein dynamics. To design proteins that function like natural ones, how do we predict their dynamics? Hannah Wayment-Steele and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1! 🧵







🚀 Excited to release BoltzDesign1! ✨ Now with LogMD-based trajectory visualization. 🔗 Demo: rcsb.ai/ff9c2b1ee8 Feedback & collabs welcome! 🙌 🔗: GitHub: github.com/yehlincho/Bolt… 🔗: Colab: colab.research.google.com/github/yehlinc… Sergey Ovchinnikov Martin Pacesa









