Kihara Laboratory
@kiharalab
Bioinformatics, protein modeling, cryoEM, drug screening, function prediction. Daisuke Kihara, professor of Biol/CS, Purdue U. YouTube: alturl.com/gxvah
ID: 271156299
http://kiharalab.org 23-03-2011 23:49:17
1,1K Tweet
2,2K Followers
2,2K Following











Happy to share that I have successfully defended my MS thesis at Purdue University! “Assessing the Accuracy of Protein Function Prediction using Machine Learning” Grateful to Dr. Kihara and everyone in the lab Kihara Laboratory


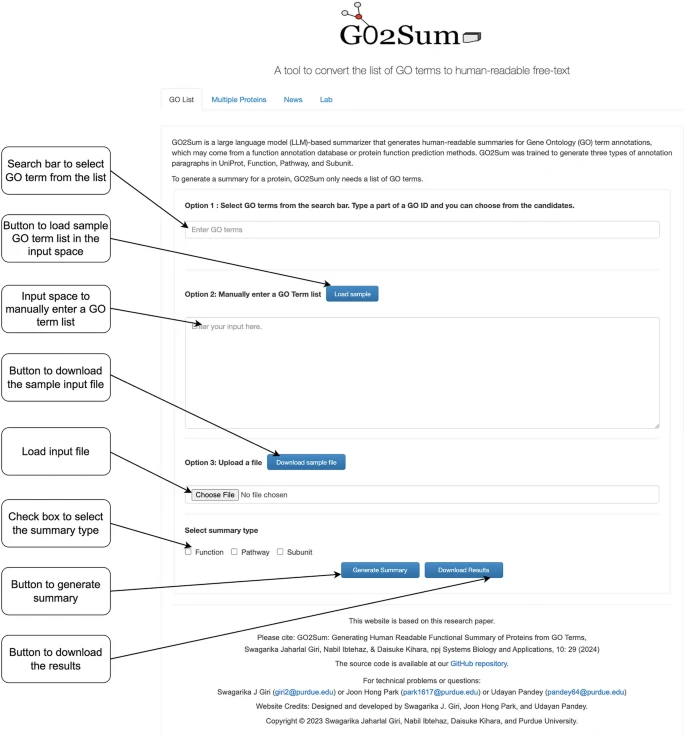
New book chapter online: Translating a GO Term List to Human Readable Function Description Using GO2Sum by Swagarika Jaharlal Giri Udayan Pandey, joonhongpark & Daisuke Kihara Methods in Mol. Biol. The server is available kiharalab.org/go2sum_server/ link.springer.com/protocol/10.10…


Dr. Eman Alnabati who did her PhD in our lab at Purdue Computer Science is featured by the Saudi Press Agency in a short video. She is now a postdoc at MIT. Great work, Eman!

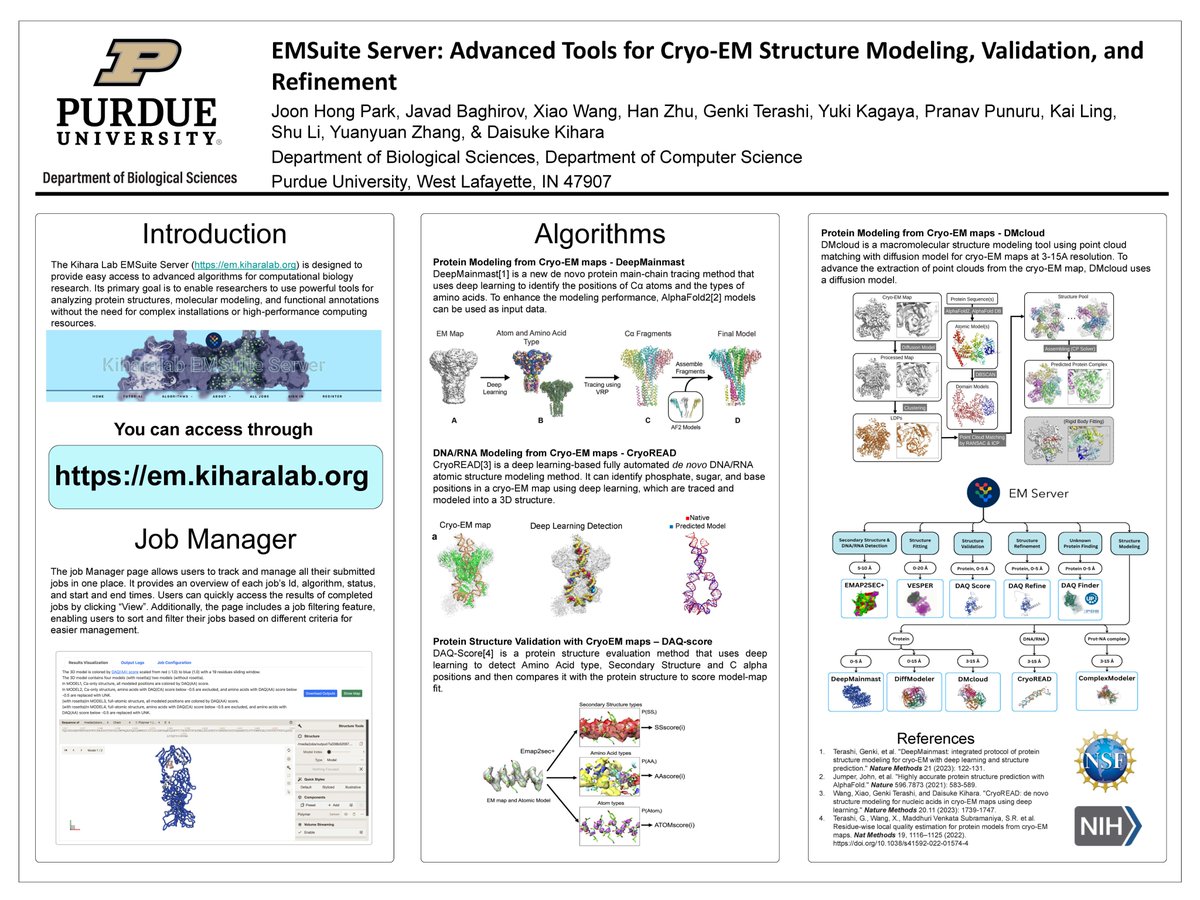
Joon Hong Park from Purdue Computer Science at American Crystallographic Association (ACA) 2025 meeting. presenting his poster about our cryo-EM structure modeling server. Visit the server for easy and accurate modeling at em.kiharalab.org


New paper accepted: "Structure Modeling Protocols for Protein Multimer and RNA in CASP16 with Enhanced MSAs, Model Ranking, and Deep Learning" Yuki Kagaya et al., Proteins. This reports our strategies in CASP. The initial submission available at ggnpreprints.authorea.com/doi/full/10.22…