
Judit Villen
@juditvr
Proteomics, mass spectrometry, systems biology
Professor at UW Genome Sciences
Mother, runner, outdoor enthusiast
ID: 319393041
18-06-2011 01:38:48
55 Tweet
818 Followers
88 Following

New preprint from the lab Miguel Martin Perez in collaboration with the Kaeberlein lab Matt Kaeberlein: Protein kinase C is a key target for attenuation of Leigh syndrome by rapamycin biorxiv.org/content/10.110…


Thesaurus, from Judit Villen and colleagues: a new proteomics search engine that finds phosphopeptide positional isomers from PRM and DIA mass spec data. nature.com/articles/s4159…

New automated end‐to‐end phosphoproteomic sample preparation method for multidimensional #CellSignaling studies --> bit.ly/2Mf31LL from Judit Villen UW Genome Sciences #proteomics #phosphoproteomics #MSBmethod


01/2020 Issue --> embopress.org/toc/17444292/2… The cover (by SciStories LLC) highlights two automated sample preparation methods for #phosphoproteomics -R2-P2 from Judit Villen UW Genome Sciences- & #clinicalproteomics -autoSP3 from J Krijgsveld DKFZ- #proteomics





Excited to share our review on PTM crosstalk Molecular & Cellular Proteomics with Sam Entwisle Judit Villen ! We discuss the basic modules of PTM crosstalk and focus on emerging technologies to study PTM crosstalk proteome-wide! #Proteomics #TeamMassSpec doi.org/10.1016/j.mcpr…


I am missing #ASMS2021 this year. But several cool presentations from students and postdocs in my lab on phosphoproteomics, MS acquisition methods, and functional annotation of protein variants Anthony Barente Mario Leutert Alexander Hogrebe, PhD Ian Smith @noncanonikyle


Happy to share new preprint from superstar postdoc Mario Leutert in my lab. One of a kind phosphoproteomic study (101 perturbations x 6) enabling cool analyses and novel discoveries in signaling. The regulatory landscape of the yeast phosphoproteome biorxiv.org/content/10.110…

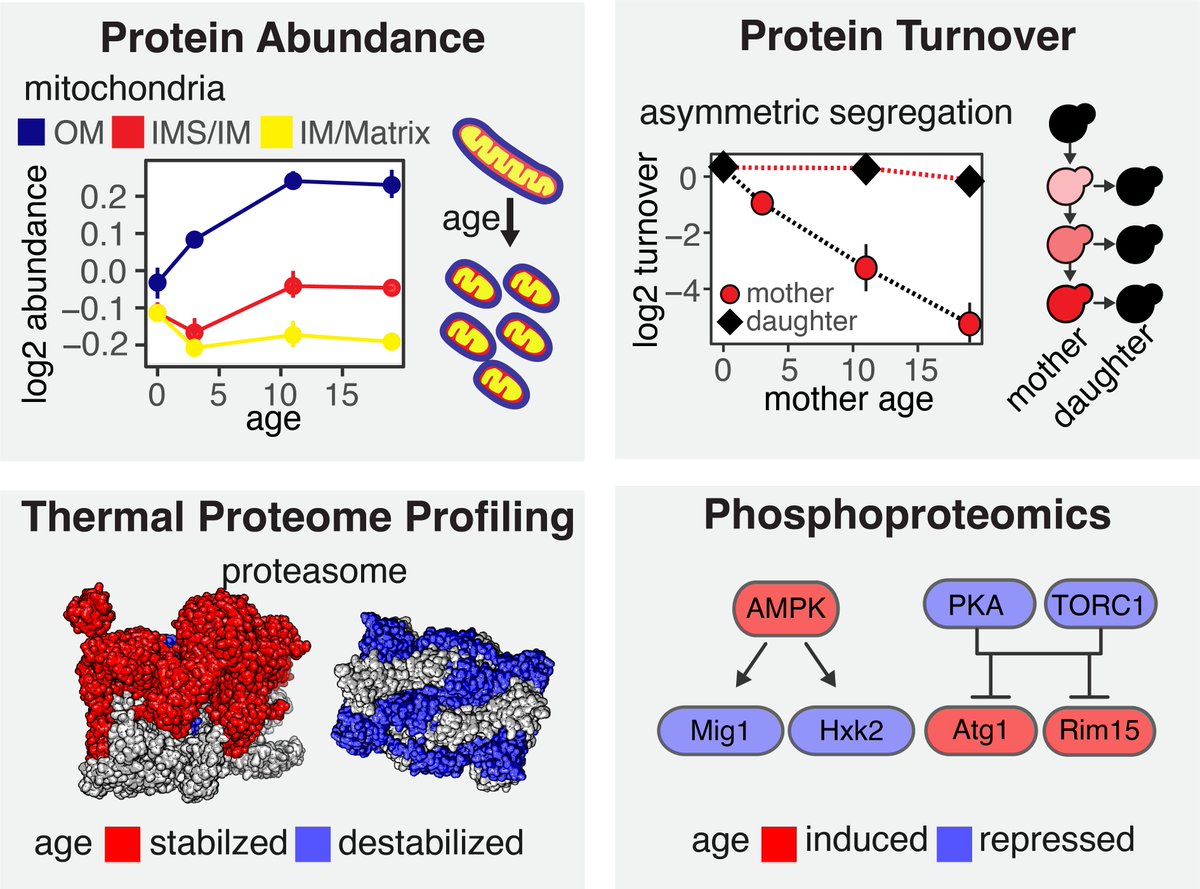
Excited to share our newest #preprint: Multidimensional proteomics identifies molecular trajectories of cellular aging and rejuvenation. A great project with the Judit Villen Maitreya Dunham Matt Kaeberlein teams! 📄Read: biorxiv.org/content/10.110… 💾Explore: rlsproteomics.gs.washington.edu 1/9


Very proud of fearless graduate student Alexis Chang for facilitating HT phosphotyrosine studies through this method.

