
Joshua Hamey
@joshuahamey
Post-doctoral researcher at UNSW using mass spectrometry to study protein post-translational modifications.
ID: 1382574859692118016
15-04-2021 06:01:55
25 Tweet
40 Followers
44 Following

Our systematic investigation into how the epigenetic regulator PRMT6 recognises its substrates - just published in The FEBS Journal. Thanks to the Bauer and Kolb labs at Philipps‐University Marburg for this fantastic collaboration. febs.onlinelibrary.wiley.com/doi/10.1111/fe…

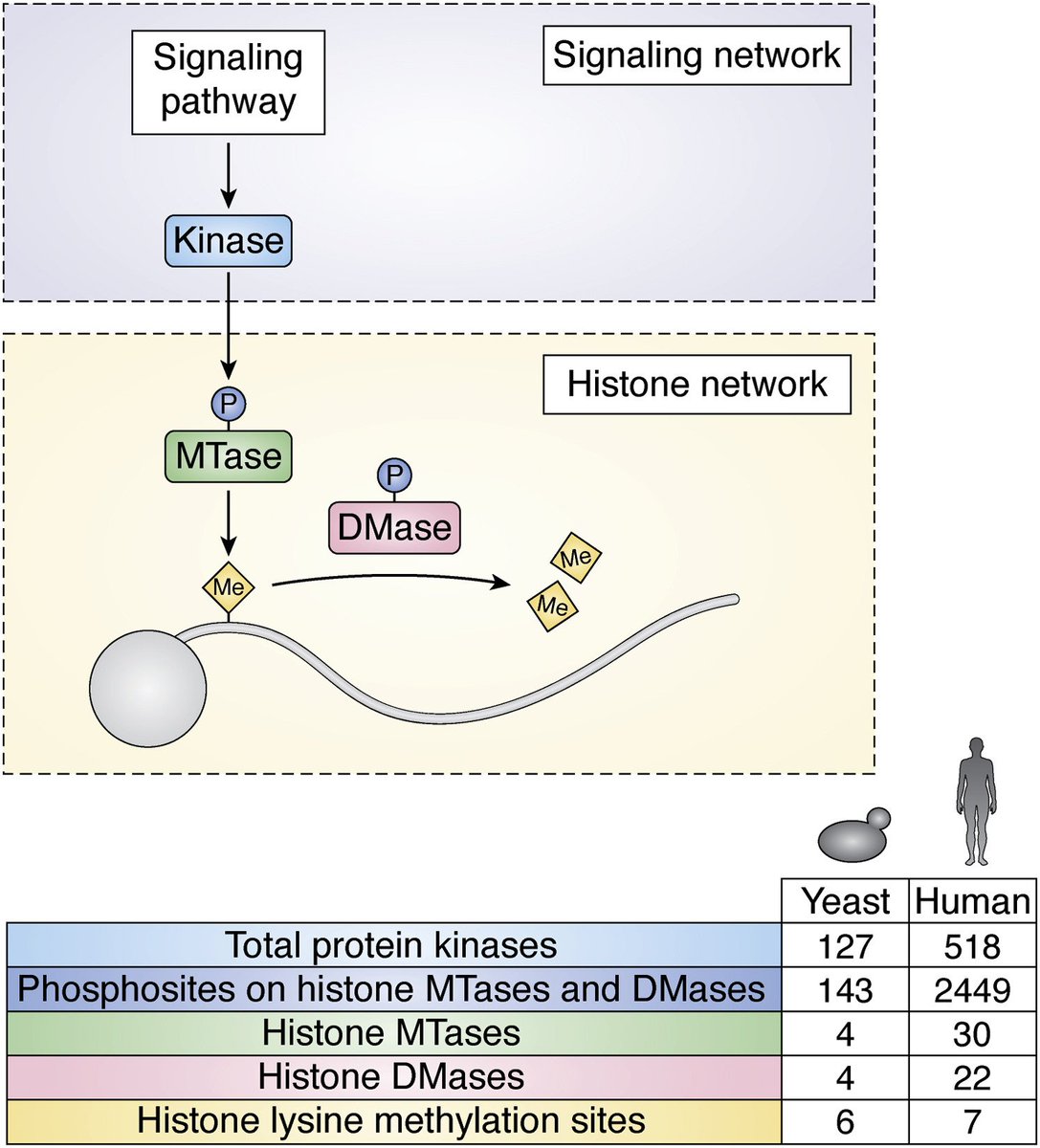
Our study discovering new phosphorylation and methylation events in the yeast proteome, including instances where they co-occur on the same protein molecule - out now in Journal of Proteome Research. pubs.acs.org/doi/full/10.10…

Do you have a budding interest in epigenetics, chromatin biology, or histone modifications? Check out our review Journal of Biological Chemistry where we cover everything you need to know about histone methylation and how it’s regulated in our lab’s favourite model eukaryote. sciencedirect.com/science/articl…

Separovich and Wilkins highlight sites of lysine methylation in the context of their biological roles and the domain architecture and structural features of methyltransferase and demethylase enzymes — UNSW Biotechnology & Biomolecular Sciences UNSW #chromatin #epigenetics jbc.org/article/S0021-…


Systematic investigation of PRMT6 substrate recognition reveals broad specificity with a preference for an RG motif or basic and bulky residues buff.ly/3AgShmg By Joshua Hamey and colleagues UNSW Philipps-Universität Macquarie University #PRMT6 #histonemethylation #substratespecificity


Just in the nick of time for my upcoming thesis submission, our manuscript has been published as an Article in Press Journal of Molecular Biology! A big thanks to everyone who contributed to this work Joshua Hamey Tara Bartolec sciencedirect.com/science/articl…

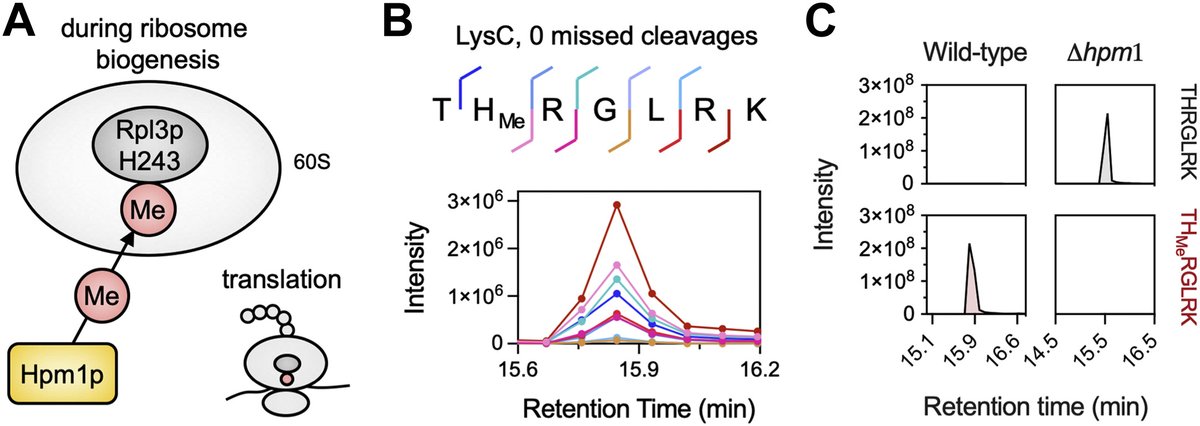
Super thrilled our new paper is out now Molecular & Cellular Proteomics ! We use qXL-MS to compare interactomes & structural proteomes between yeast strains & show how it can uniquely mechanistically explain protein function which is invisible to abundance analysis alone! mcponline.org/article/S1535-…
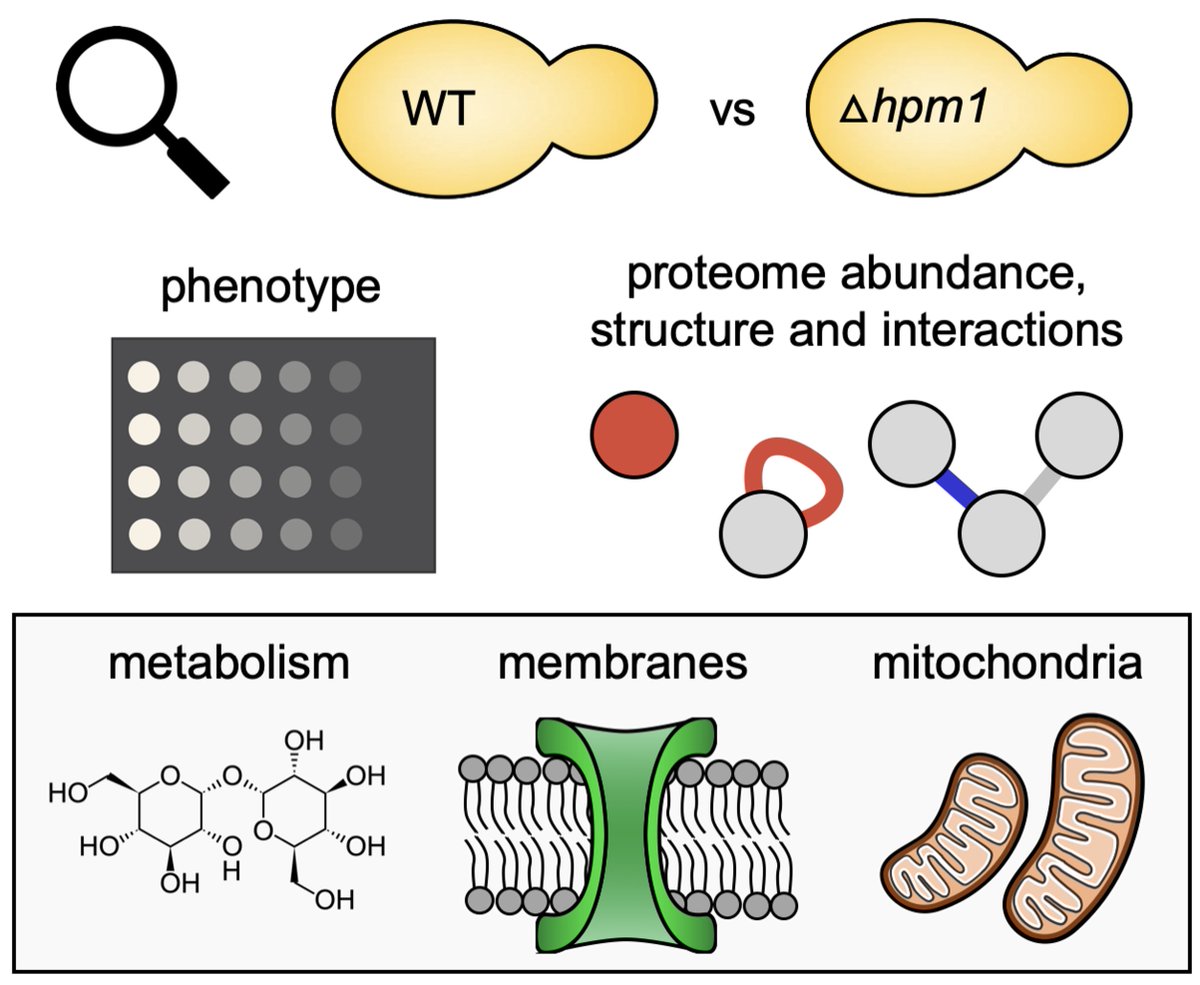



Methyltransferase Hpm1p is the only known histidine methyltransferase in Saccharomyces cerevisiae. In this Molecular & Cellular Proteomics paper, researchers investigated how the loss of Hpm1p generates different phenotypes. mcponline.org/article/S1535-…

Join us on 30 Aug at 12:30pm AEST for two exciting talks hosted by UTS Science Australian Institute for Microbiology & Infection Joshua Hamey will present his UNSW Biotechnology & Biomolecular Sciences work on #methylation in yeast David Nipperess will present his work on expected survival as an indicator of biodiversity zoom:utsmeet.zoom.us/j/89426903922








