
Jin Sun @OUC
@jinsun_hkust
Deep-sea biology, Malacologist, Genomics, Chemosymbiosis, Phylogeny
ID: 813304847038443520
https://iemb.ouc.edu.cn/hyhnhcstxtsys/list.htm 26-12-2016 08:46:00
844 Tweet
306 Takipçi
325 Takip Edilen

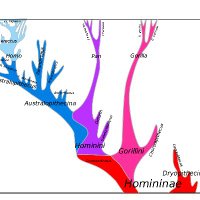
CAnDI: a new tool to investigate conflict in homologous gene trees and explain convergent trait evolution academic.oup.com/sysbio/advance… Society of Systematic Biologists






Glad to see this paper finally out! An episodic burst of massive genomic rearrangements and the origin of non-marine annelids NatureEcoEvo Institute of Evolutionary Biology (IBE) Metazoa Phylogenomics Lab CSIC European Research Council (ERC) nature.com/articles/s4155…






torchtree: flexible phylogenetic model development and inference using PyTorch academic.oup.com/sysbio/advance… Society of Systematic Biologists





