
Jesper V. Olsen
@jespervolsen
ID:756899969995595780
23-07-2016 17:13:10
24 Tweets
1,4K Followers
1,2K Following



Hey #Proteomics enthusiasts, come and join us next summer in Brixen. Bavarian Center for Biomolecular Mass Spectrometry part of orga team of this great event. More info will follow soon… #TeamMassSpec #Bioinformatics

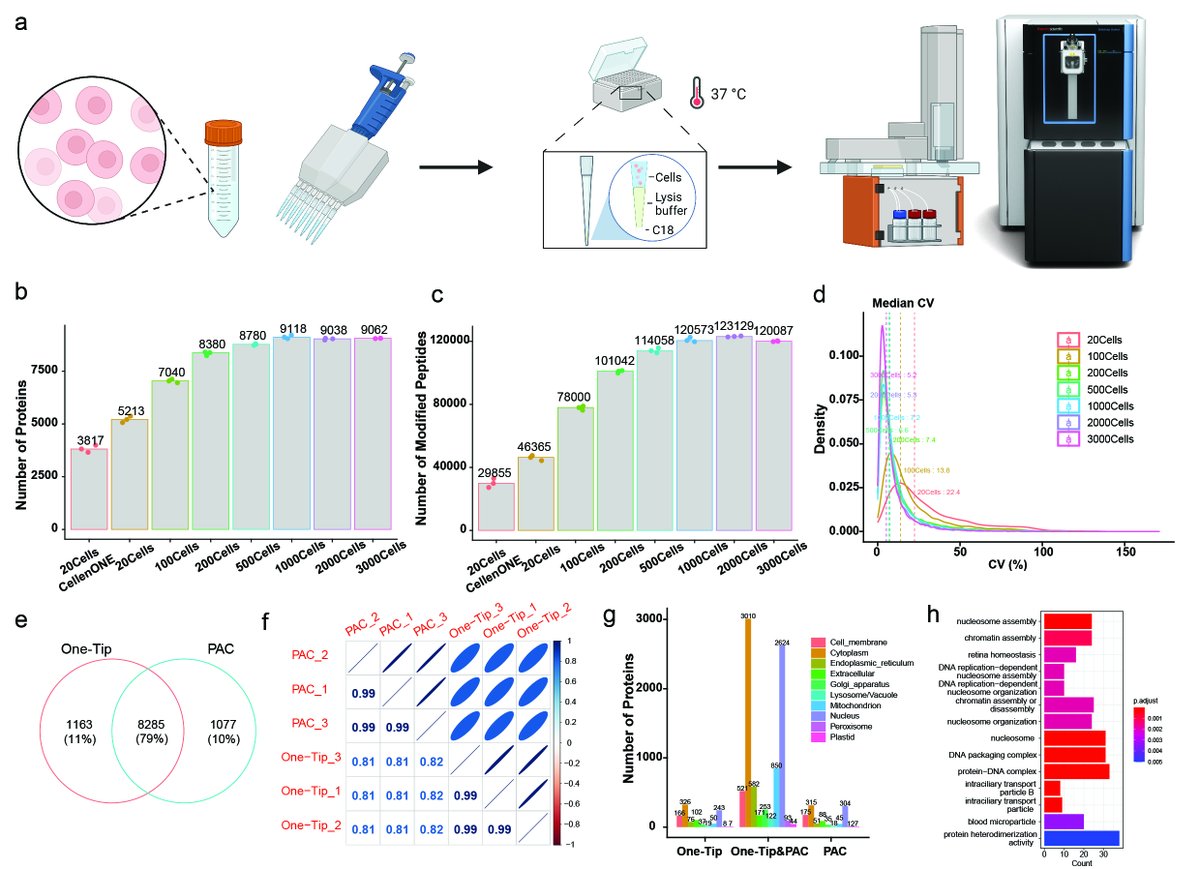
Fully automated workflow for integrated sample digestion and Evotip loading enabling high-throughput clinical proteomics biorxiv.org/cgi/content/sh… #biorxiv_molbio

🚀 Here is Jesper V. Olsen from CPRatUCPH for the #proteomics session at #proteinsignaling2023
🗣️ The talk is about #SingleCell proteomics with narrow-window DIA provides high #proteome coverage
#science #research
Novo Nordisk Foundation



🔬 Exciting new research on EGFR from JesperOlsenLab ! Akihiro Eguchi, Kristina B. Emdal and colleagues used high-throughput proteomics to uncover the molecular mechanisms behind EGFR's response to its 6 highest-affinity ligands. Check out the preprint: biorxiv.org/content/10.110…

Less is more with the One-Tip method! From single mouse zygotes to HeLa cells, discover unmatched proteome depth & efficiency in our new preprint from Zilu Ye Pierre Sabatier , coupling Evosep and #Orbitrap #Astral #TeamMassSpec . shorturl.at/ciGY6

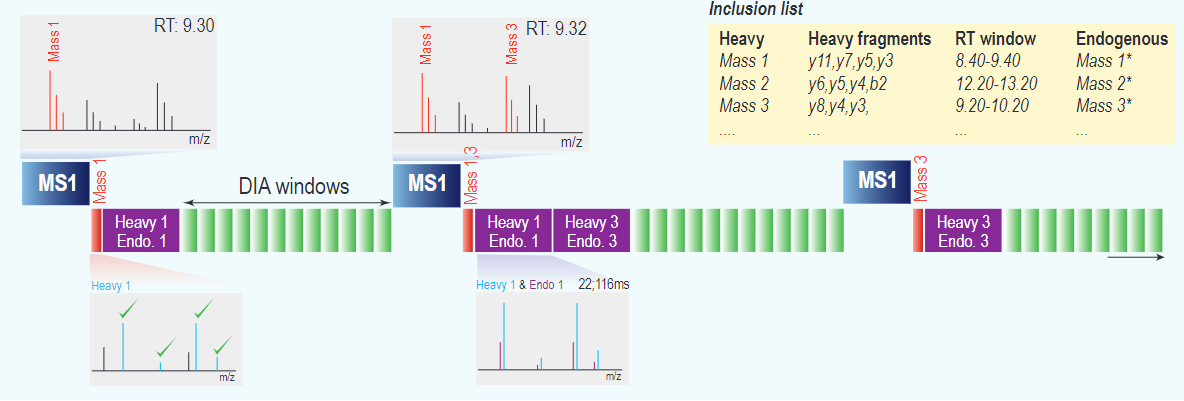
Our work at JesperOlsenLab on describing and benchmarking hybrid-DIA methodology for simultaneous targeted and discovery phosphoproteomics is now out in Nature Communications: nature.com/articles/s4146…

Exciting developments in #proteomics with the new ultra-fast scanning DIA strategy in Orbitrap Astral MS. This innovation is set to reshape proteome analysis, blending speed, precision, and accuracy. Dive into the details at bioRxiv: biorxiv.org/content/10.110…

Boost your analysis with our new hybrid-DIA method, developed by our superstar postdoc Ana Martinez del Val, that allows to combine targeted and discovery #proteomics in just one run. We have used it to boost drug screening phospho-research on spheroids. Check it at biorxiv.org/cgi/content/sh…

Proud to be named a Highly Cited Researcher 2022 by Clarivate for Academia & Government. See the full list bit.ly/3fPhNLo #HighlyCited2022

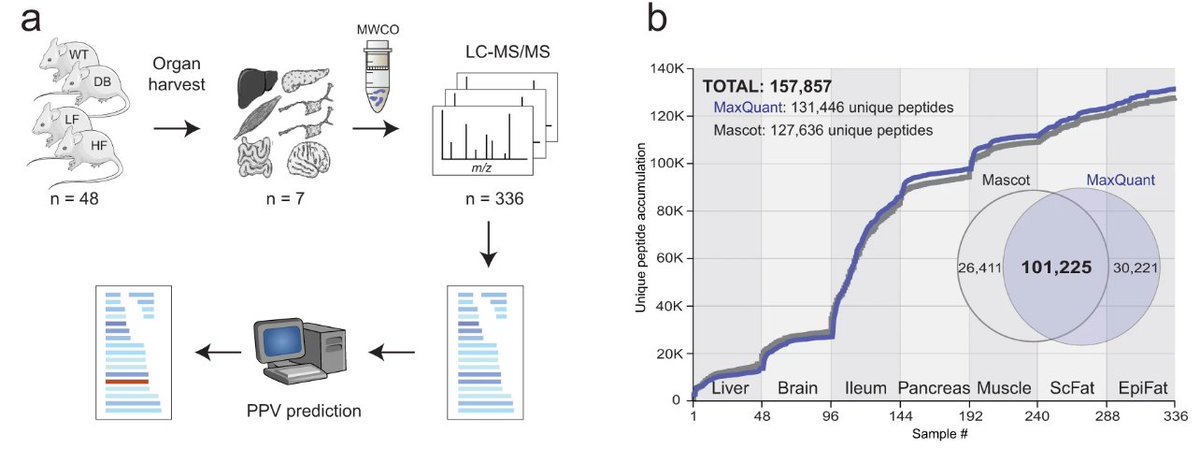
New cool collaboration paper in Nature Communications with researchers at Novo Nordisk: rdcu.be/cXXCa
We developed a powerful machine learning method to identify new bioactive peptides from large-scale #peptidomics data collected from seven different mice organs.


Proud to be named a Highly Cited Researcher 2021 by Clarivate for Academia & Government. See the full list bit.ly/3qEBjxg #HighlyCited2021

Great line up at the exclusive #ProteinSignaling Conference this November 14th near Copenhagen. Check it out and apply with an abstract by tomorrow. CPRatUCPH, Novo Nordisk Foundation
cph-bioscience.com/en/events/prot…





