Jean Loup Faulon
@jeanloupfaulon
Research Director @Inra_France
Professor @OfficialUoM
ID: 1958786215
https://www.jfaulon.com/ 13-10-2013 14:02:23
25 Tweet
394 Followers
283 Following

Olivier Borkowski presents his cool work demonstrating the use of active learning in cell free system optimization #cellfreesystems19



A bit delayed on PMID:32312991… nice collaborative work between Olivier Borkowski and Mathilde Koch with the help of Amir Pandi, Soudier Paul, Agnès Zettor, and Angelo Batista, congrats!


Happy to announce an open research associate position in my lab BioRetroSynth Research Group for biosensor engineering. Information on how to apply can be found at jfaulon.com/open-positions/



Happy to share #GalaxySynBioCAD our latest publication Nature Communications (rdcu.be/cUzbo) a first step toward a user-friendly operating system for automated metabolic engineering and Synbio. Can be freely used at galaxy-synbiocad.org
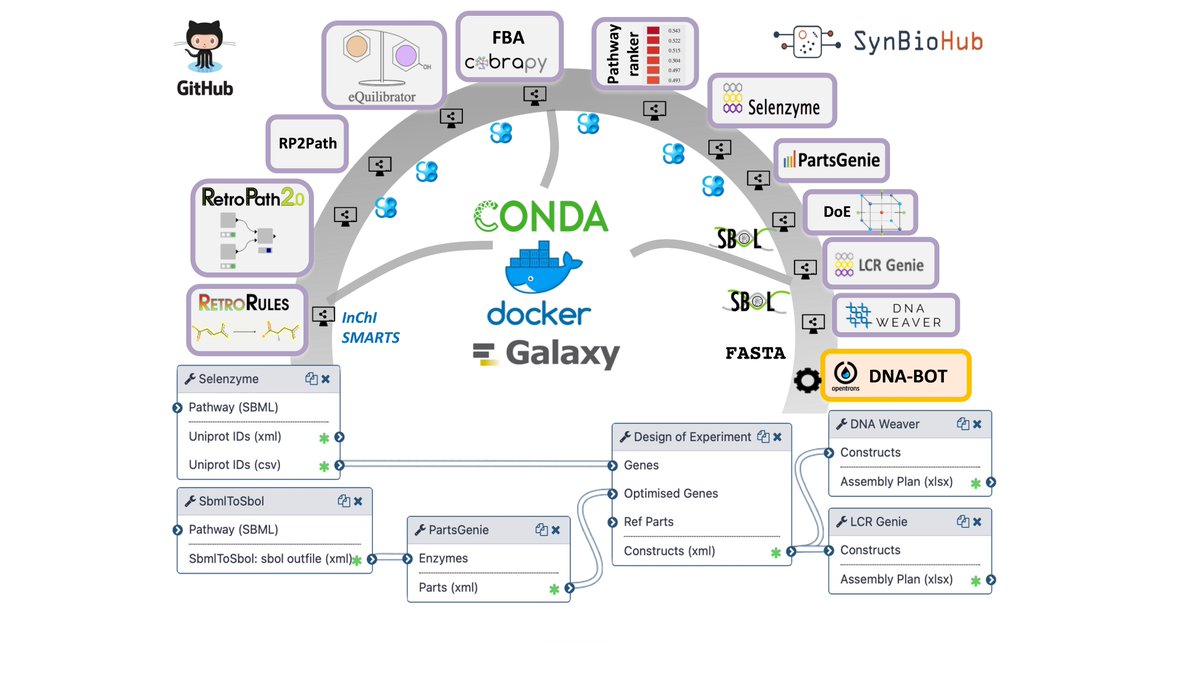

Check out the genesis and feedback about the development of the #GalaxySynBioCAD portal with our "behind the paper" blog post: "Towards automated synthetic biology” (go.nature.com/3qwIqWV). Thanks to Joan and Thomas BioRetroSynth Research Group

Delighted to announce the 2023 Turing Workshop on AI, Engineering Biology & Beyond. Come join us in Edinburgh! 13-14 March 2023 tinyurl.com/5xh9x55k Organised by Edinburgh, Bristol and UCL with Thomas Gorochowski & CSSB Lab Centre for Engineering Biology ANC@Edinburgh School of Informatics, The University of Edinburgh The Alan Turing Institute #synbio #AI


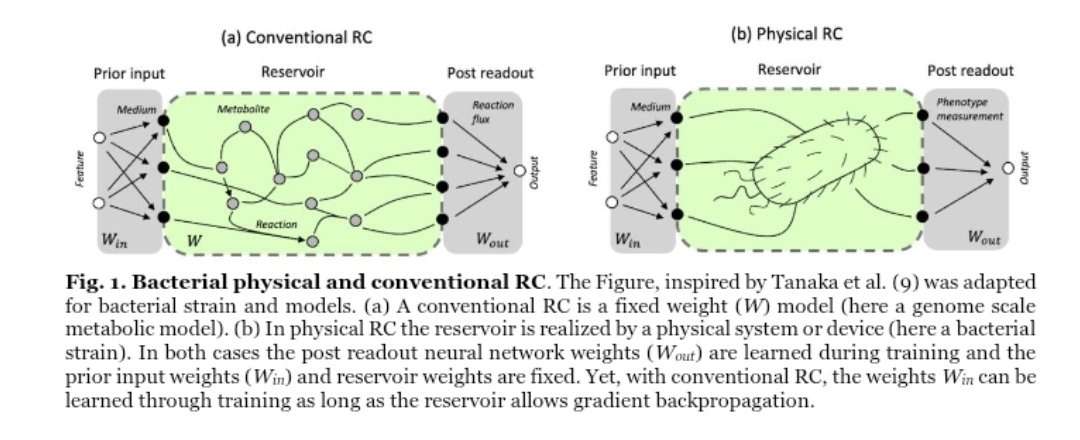
Happy to announce the release of our latest work published Nature Communications. We integrated machine learning with mechanistic modeling to improve microorganisms‘ genome-scale phenotype predictions doi.org/10.1038/s41467…






Cellular computing without bioengineering by Jean Loup Faulon lab. doi.org/10.1101/2024.0…