Yihao Zhang
@tomaszhang4482
MD/PhD. Transcription and translation regulation, molecular oncology, bioinformatics
ID: 798150511232811009
14-11-2016 13:08:05
71 Tweet
1 Takipçi
39 Takip Edilen









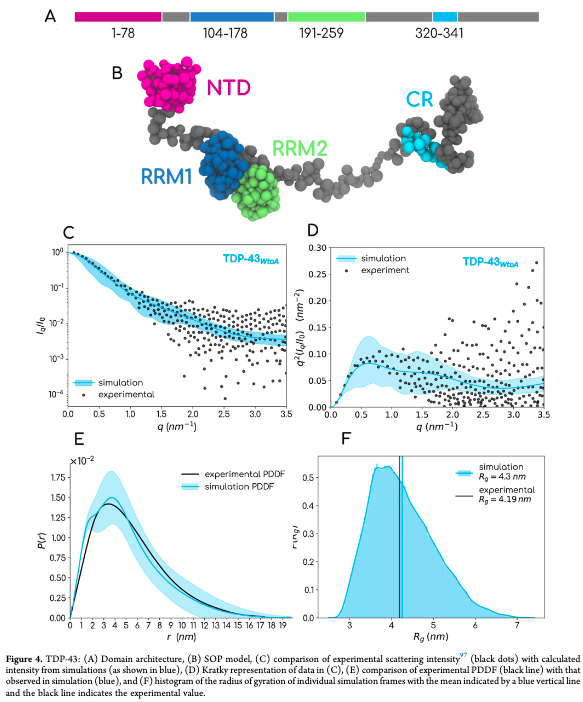
pubs.acs.org/doi/full/10.10… Very chuffed to see this out in JCIM & JCTC Journals!! CGFF of full length FUS, hnRNPA1, TDP-43, G3BP1, TIA-1, Poly UBQ, HIV GAG and more. Wonderful experience working with KrishnaKanth B . Thanks Kresten Lindorff-Larsen Mittal Group @TAMU Debayan Chakraborty for discussions.






DrNadaSalma⚕ source: scribd.com/document/76905…