Dave Moi
@steg0s4urus
🔬 Postdoctoral Researcher working at the @dbc_unil for the @DessimozLab | Bridging Borders | #Bioinformatics & #MachineLearning 🤖 |
ID: 851769488
28-09-2012 21:28:45
1,1K Tweet
243 Takipçi
324 Takip Edilen


Evo: a foundation model for genomics! Using a well suited architecture, Evo learns from billions of bp of genomic sequence and performs well on several zero-shot prediction tasks on RNA, DNA and protein. Beautiful paper from Brian Hie & Patrick Hsu at Arc Institute


New open access paper out in NatureEcoEvo TLDR: - Humans pass on more viruses to other vertebrates than they do to us - Generalist viruses (with broad host range) require fewer mutations to infect a novel host 1/ nature.com/articles/s4155…

nature.com/articles/s4155… Have a look at the article I reposted by Prof Francois Balloux, Cedric C.S. Tan and Lucy van Dorp if you're interested in viruses and their evolutionary signature when jump hosts. I had the pleasure of writing a news and views piece about this work with Christophe Dessimoz


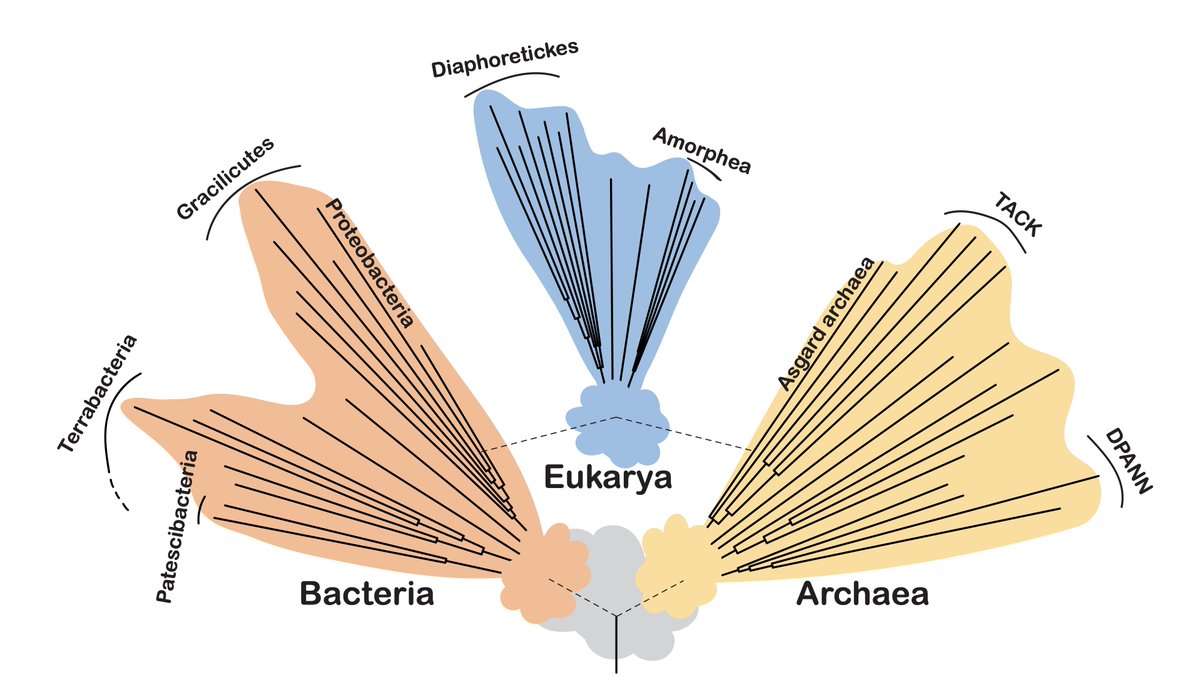
🧬🔬 Nice dive into how #metagenomics & single-cell genomics are revolutionizing our understanding of the tree of life by @eme_laura & Daniel Tamarit 🧬 #MicrobialDiversity #genomics #singlecell #biodiversity #protists




7/ In particular, we showcase gLM2's ability to directly learn coevolutionary signal in protein-protein interfaces with no supervision! The learned contact maps can be extracted using Zhidian Zhang et al's categorial Jacobian method.





This is a foundational achievement. MMseqs2-GPU virtually *erases* the bottleneck of homology search in many workflows, including structure prediction. Christian Dallago @milot_mirdita @thesteinegger, Felix, Bertil, and Alejandro too! developer.nvidia.com/blog/boost-alp…




Fantastic #Sinergia retreat on #animalorigins! A day packed with insights into #multicellularity, #ichthyosporeans, #choanoflagellates & more—capped with a Swiss-style finale. Huge thanks to 𝘋𝘪𝘳𝘬 Stefano Pascarelli Dave Moi Christophe Dessimoz Pedro Beltrao 🚺

Dessimoz Lab representation at #ISMB2025 * Phylogenies with protein structures, #FoldTree - Dave Moi * Ancestral genome reconstruction: #EdgeHOG - Charles Bernard * Gene duplications and functional evolution Alex WV and Irene Julca