
Sophia J. Wagner
@sophiajwagner
PhD student @helmholtz_ai and @TU_Muenchen. AI for Computational Pathology
ID: 1383872513491365892
18-04-2021 20:01:01
34 Tweet
303 Takipçi
207 Takip Edilen

Great success at our parallel Session 2b: "Applied AI" today! Thanks to our outstanding speakers Annika Bande, Yunfei Huang, Sophia J. Wagner for showcasing the infinite applications of AI 🙌 #HelmholtzAICon #AppliedAI


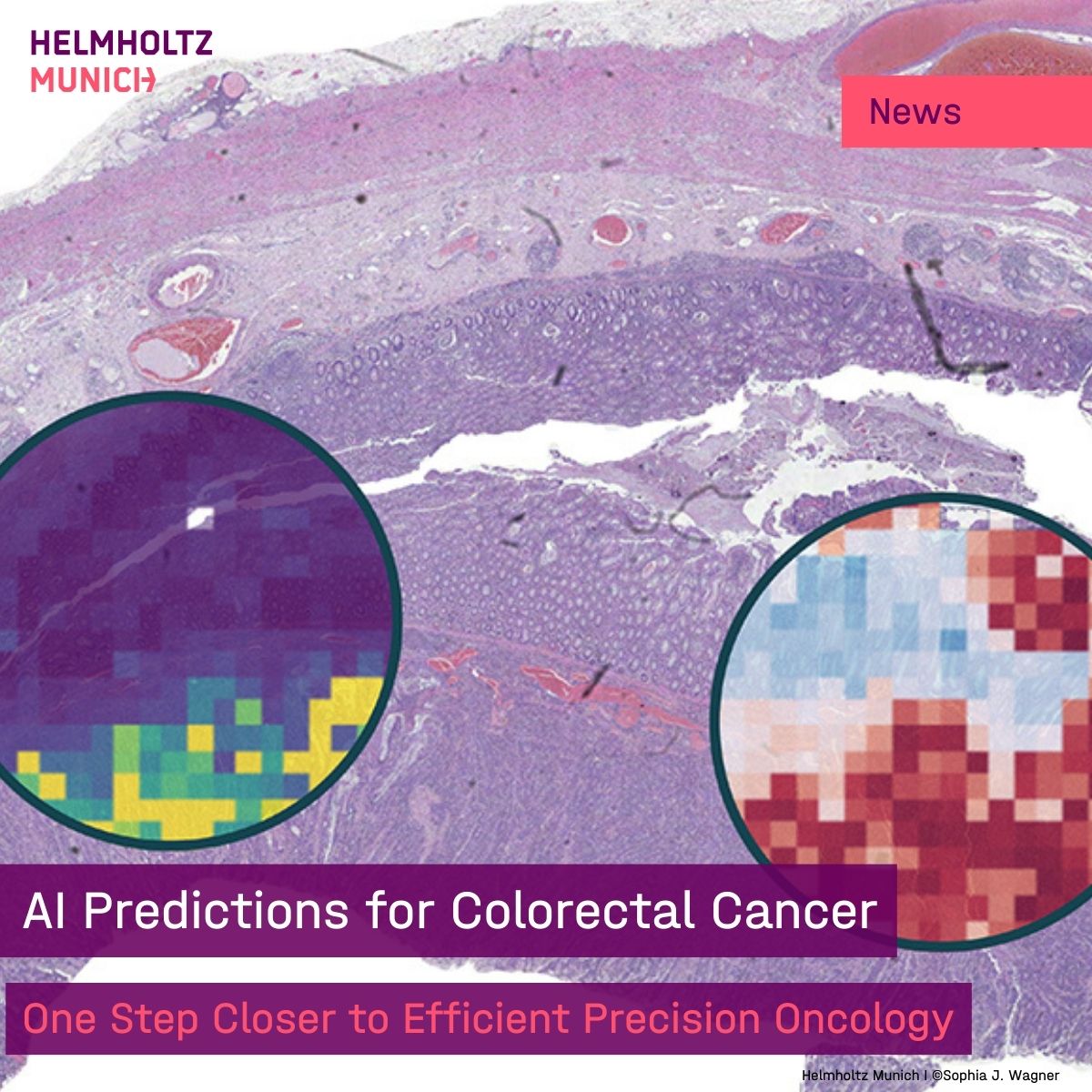
📝Hope for Colorectal #Cancer Using #AI, scientists from #HelmholtzMunich & TU Dresden boost #precision #medicine in #oncology: Novel model for #biomarker detection helps to analyze tissue quicker, speeding up treatment decisions. 👉more insights: t1p.de/pmcs0 Jakob Nikolas Kather

🚀 FIRST-EVER Marr & Peng Lab Hackathon in beautiful Oberammergau! 👩💻 Was a great time to kickstart some promising projects 🥳 thanks to all the participants, especially co-organizers Benedikt Mairhörmann Rushin Gindra and of course carsten marr and Tingying Peng for making it all possible!


Happy to share DinoBloom 🦖🌺 for generalizable cell embeddings of blood🩸and bone marrow💉smears in #hematology developed w Valentin Koch, take a look at this thread👀 preprint: arxiv.org/abs/2404.05022 github: github.com/marrlab/DinoBl… Helmholtz Munich | @HelmholtzMunich carsten marr Tingying Peng

I am tremendously excited to announce that my work on 3D computational pathology has been published in Cell !! 🥰🎉 This has been truly a wonderful journey for the past few years. I really want to thank my wonderful mentors Professors Faisal Mahmood and Jonathan Liu



⚡️📣Delighted to announce MMP, a prototype-based multimodal framework combining histology and transcriptomics for cancer outcome prediction, to appear in #ICML 2024 ICML Conference. Congratulations to our superstar postdoc Andrew H. Song and rest of the team who helped the study.


1/ 🧵Introducing UNICORN,🦄 the first #AI model for integrating multi-stain #histopathology images for classification, designed to tackle missing data during both training and inference. Great colab between Helmholtz Munich | @HelmholtzMunich & TU München German Heart Centre! the architecture ⬇️


Join us at our poster T64 at #MICCAI2024 to talk about DinoBloom 🦖🌺 the first feature extractor in hematology for bloodsmears🩸 All models are available at github.com/marrlab/DinoBl… Developed w Valentin Koch, supervised by Tingying Peng carsten marr Helmholtz Munich | @HelmholtzMunich TU München


Excited to share TITAN, a multimodal foundation model for pathology trained on >330k WSIs, synthetic captions, and reports🔬⚡ co-lead w Tong Ding Andrew H. Song Richard J. Chen Thanks to Faisal Mahmood Tingying Peng! Preprint: arxiv.org/abs/2411.19666 Model: huggingface.co/MahmoodLab/TIT…




