Single Cell Omics Germany (SCOG)
@singlecellomics
Connecting researchers using & developing single cell technologies. Providing a collaborative platform, organizing workshops & hackathons, exchanging expertise.
ID: 1021858152134914049
https://linktr.ee/singlecellomics 24-07-2018 20:42:23
2,2K Tweet
4,4K Takipçi
596 Takip Edilen

📢 Starting now! Don't miss Céline's lecture "Mechanisms of cell plasticity in breast cancer". Céline Vallot, Institut Curie, #SCOGVLS ✏️ Register: bit.ly/scogvls

We thank the speakers, organizers and ca. 200 participants of the 16th Berlin Summer Meeting 'The MDC Celebrates 15 Years of BIMSB', 6-7 Sept 2023, for 2 days of superb presentations & discussions on the latest exciting findings in systems biology! Max Delbrück Center #mdcBIMSB2023



Our journey of multiplexed DIA, encompassing applications in spatial #singlecell #proteomics, is showcased on the cover of the latest issue of Mol Syst Biol. We are excited about the prospects of sensitivity, reproducibility and high-throughput applications in clinical proteomics.

We are thrilled to share our latest work, a collaborative effort with Treutlein lab, in Nature (nature.com/articles/s4158…). We introduce CHOOSE, a powerful screening system within cerebral organoids, through which we identified developmental defects associated with autism. 🧵1/7



Excited that veloVI is out Nature Methods! Led by Adam Gayoso & Philipp Weiler w awesome Yosef Lab, we combine deep generative modeling with a mechanistic single-cell transcriptional model for better RNA velocity analysis with uncertainty quantification. nature.com/articles/s4159…



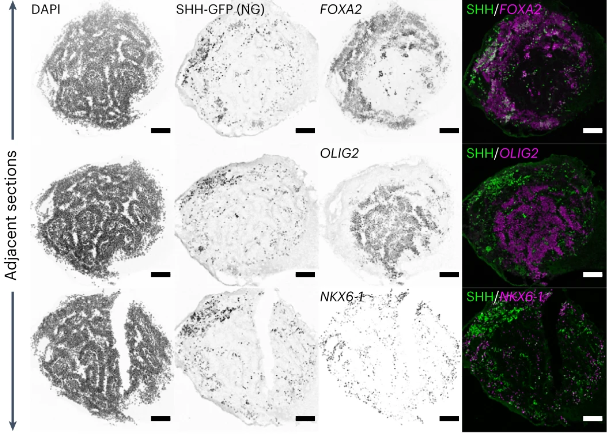
A workflow developed by Nikolaus Rajewsky, Ivano Legnini and colleagues combines optogenetic perturbations and spatial transcriptomics to enable insights into cell fates and spatiotemporal gene patterns in organoids. OA paper: nature.com/articles/s4159…





36 datasets, 26 protocols, 1.7 million harmonized cells! First edition Human Neural Organoid Cell Atlas. Important work from ZhiSong He 何志嵩 Jonas (josch1@bsky) Leander Dony Treutlein lab Fabian Theis. Strong community effort Sergiu P. Pasca Giorgia Quadrato. #organoids D-BSSE, ETH Zurich Institute of Human Biology


(1/n)scPoli is now published in Nature Methods shorturl.at/jvyL0; see below. scPoli is an open-world learner for multi-scale analysis of scRNAseq/ATAC-seq across samples and species with cell types and conditions hierarchies. With scPoli you can:


#AI accelerates & enables #therapies! 👉Learn how Prof. Fabian Theis & team at #HelmholtzMunich & TU München transform info of #BigData into medical #applications: 🔗Einzelzell-Atlas der Lunge: t1p.de/j5ogs 🔗Passionate about AI in #Health: t1p.de/jdg1v



Save the date for the 17th Berlin Summer Meeting: AI’s Adventures in Genomics, 6-7 June 2024! Speakers include Vikram Agarwal @vagar112, Rebekka Burkholz, Julien Gagneur gagneurlab, Andreas Keller Chair for Clinical Bioinformatics, Georg Seelig Georg Seelig, Ewa Szczurek & Josh Welch Josh Welch #mdcBerlin