The Sharing Scientist
@scienceshared
Bringing you the most interesting & important science news that you need, summarised & sourced directly to your feed.
Also will call out stuff others don't.
ID: 224317914
08-12-2010 18:02:14
14,14K Tweet
12,12K Takipçi
3,3K Takip Edilen













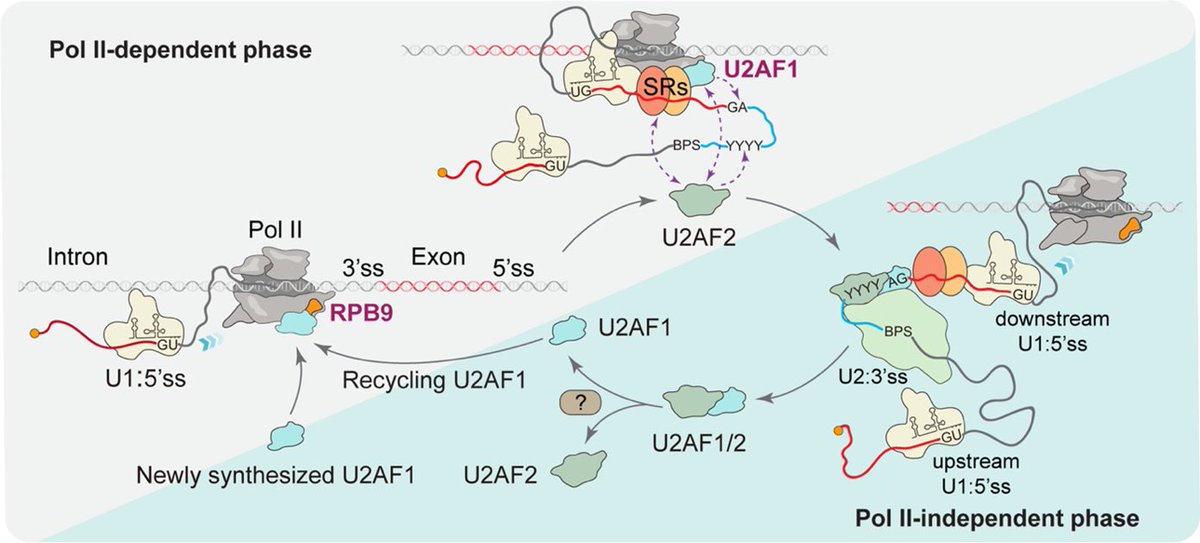
We are proud to share a paradigm-shifting publication from Westlake University in the journal Science Magazine . The study, led by Prof. Xiang-Dong Fu, reveals a fundamental mechanism of gene expression: how cells accurately identify correct splice sites during transcription.