Laurent Francioli
@lfranciol
ID: 4833103606
21-01-2016 15:12:07
101 Tweet
157 Takipçi
240 Takip Edilen


I see that the black Friday / cyber Monday sequencing deals are back this year. Just a warning if you're considering getting the Dante Labs promo: I purchased mine a year ago and still don't have anything (and their customer support couldn't care less)

We are launching the "COVID-19 host genetics initiative" Goal: aggregate genetic and clinical information on individuals affected by COVID-19 FIMM HelsinkiUni will genotype samples for FREE and make the data available to the scientific community covid-19genehostinitiative.net




📢 Only 4 days left to apply to do a PhD in my new group in Oxford! Centre for Human Genetics Nuffield Department of Medicine University of Oxford Apply here: ndm.ox.ac.uk/investigating-… Deadline: Monday (20th July) See thread below for more details 👇

Whenever I start getting too pessimistic about the US, I think about the incredible feat @BroadGenomics has managed to pull off in massive-scale COVID testing. It's exactly the kind of relentless innovation the country needs. Plus Heidi Rehm is a badass.

Interested in exploring the contribution of rare genetic variants to gene expression? Joseph Powell and I are looking for postdocs to lead analyses of a new data set of >1,000 whole-genome-sequenced individuals with scRNA-seq data. More details: garvan.wd3.myworkdayjobs.com/en-US/garvan_i…

An exciting #ASHG20 update from Genome Aggregation Database - the release of v3.1, with over 3,000 new diverse genomes added, and a bunch of new features: gnomad.broadinstitute.org/blog/2020-10-g… Massive props to the team for their hard work on this release!



Please RT! We are looking for a Scientific Operations Manager for the Novo Nordisk Foundation Center for Genomic Mechanisms of Disease Broad Institute. Come play a key role in a vibrant partnership to improve human health through genomics and data sciences: broadinstitute.wd1.myworkdayjobs.com/en-US/broad_in…



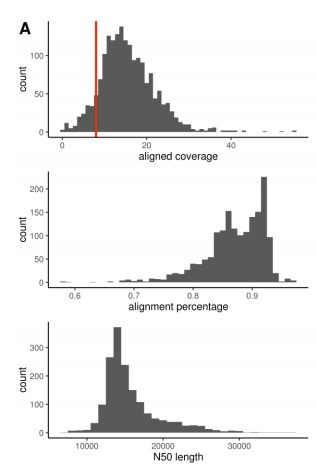
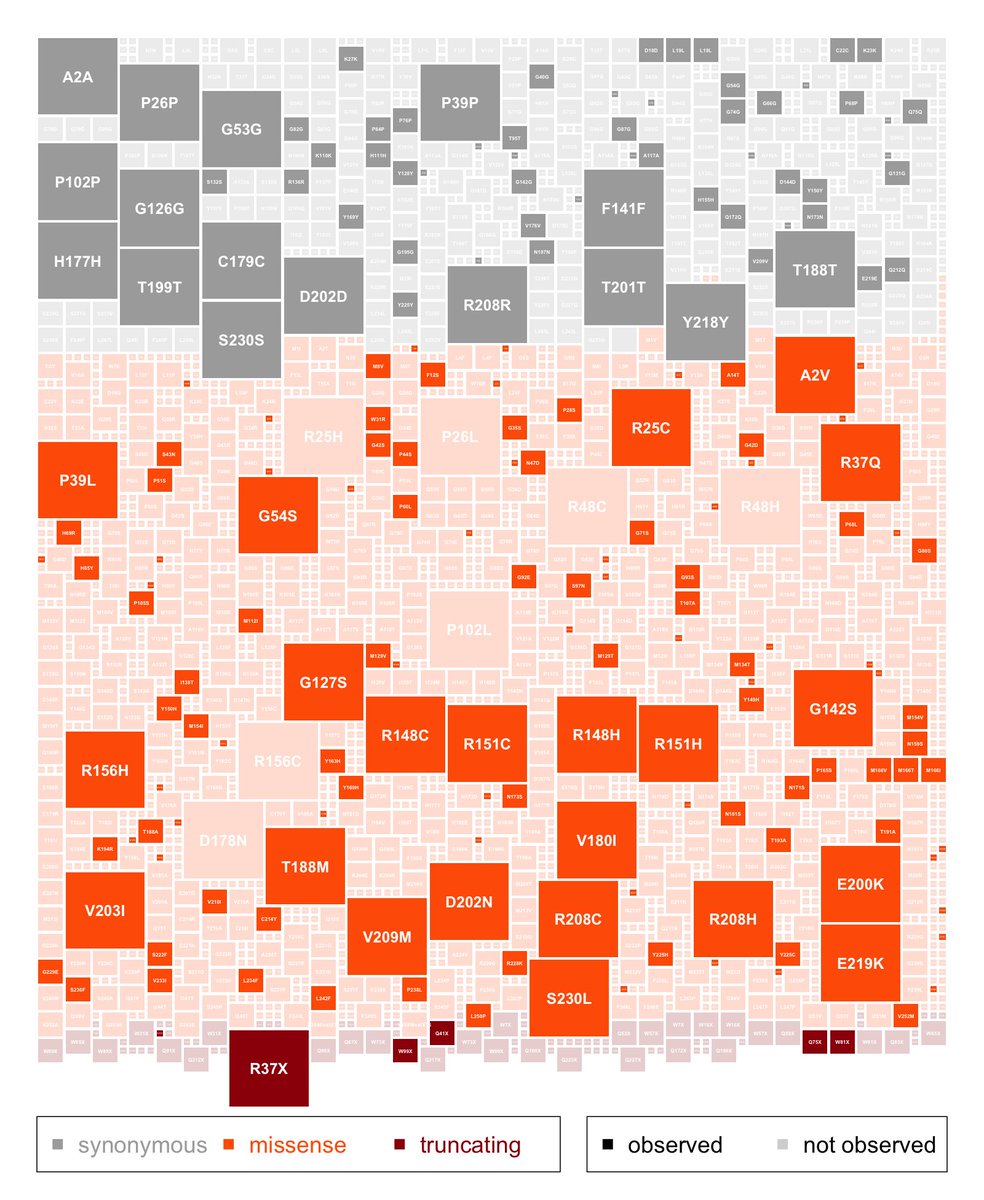
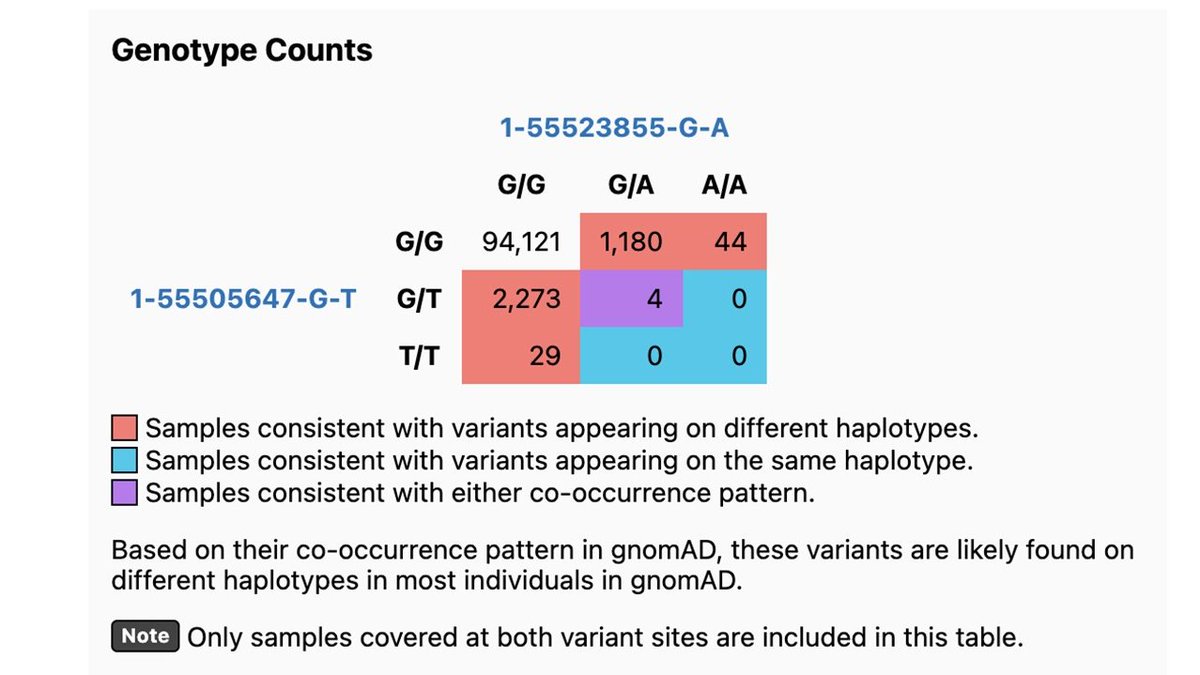
As we all continue to work on deriving the most value from WGS, we're excited to post this preprint of a genome-wide truth set for tandem repeat detection. An amazing amount of work from Ben Weisburd which has also enabled many diagnoses in our Rare Genomes Project.