Jake Robson-Tull
@jrobsontull
Doing drug discovery things. Senior Scientist at Schrodinger. Views are my own.
ID: 1248168884773048320
http://jakert.me 09-04-2020 08:40:38
228 Tweet
142 Takipçi
442 Takip Edilen



There's been several good studies on how people actually use LLMs for coding tasks. This one presents an interesting, mixed-methods, deep-dive into the topic. From Hussein Mozannar Gagan Bansal Adam Fourney and Eric Horvitz; good list of relevant cites, too! arxiv.org/abs/2210.14306



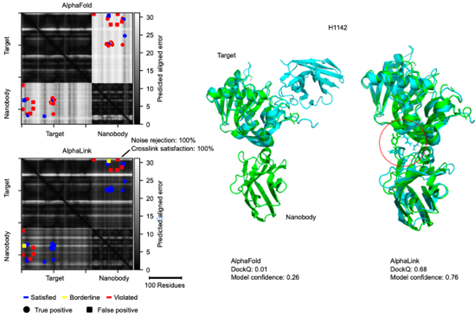
In collaboration with the RBO Lab - TU Berlin, we are pleased to present AlphaLink2 - a modified version of AlphaFold-Multimer that is capable of directly incorporating experimental residue-residue contact data from crosslinking MS (and potentially other techniques as well). (1/7)







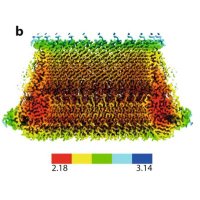

Just out from the Carvalho lab!!! Yours truly, Nik Sergejevs and colleagues together with LuLin Jiang and Matthias Trost 🇪🇺 discover a new role for the RNF185-Membralin ERAD complex in regulating Tapasin assembly with the PLC! Get it while it's hot! nature.com/articles/s4146…

Excited to share my first, first-author manuscript on the role of Signal Peptide Peptidase in membrane protein quality control. Massive thanks to our collaborators, @Marius_Lemberg Dönem Avci Robin Corey @robincorey.bsky.social and postdoc Michael van de Weijer Dunn_School (now @DunnSchool.bsky.social) journals.biologists.com/jcs/article/do…


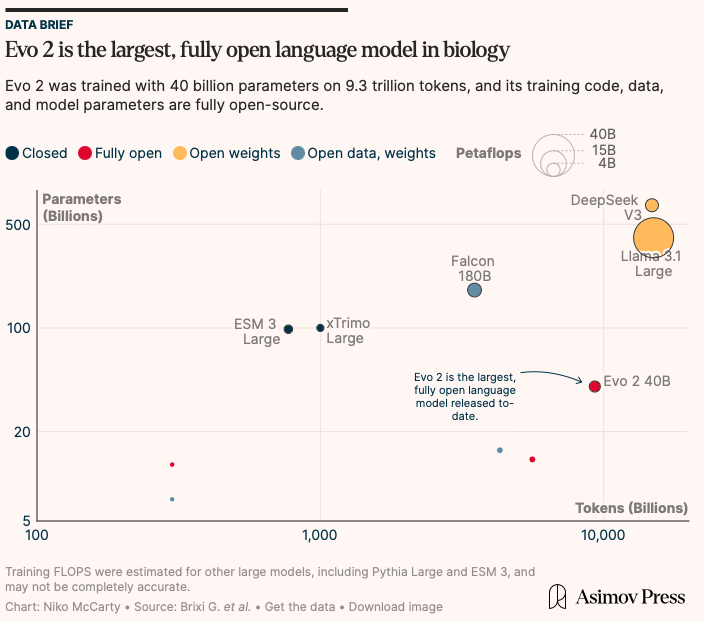
Just now: Arc Institute and NVIDIA release Evo 2, the largest AI model for biology. It is fully open-source. Evo 2 can predict which mutations in a gene are likely to be pathogenic, or even design entire eukaryotic genomes. We covered it in Asimov Press! Check it out 🔻

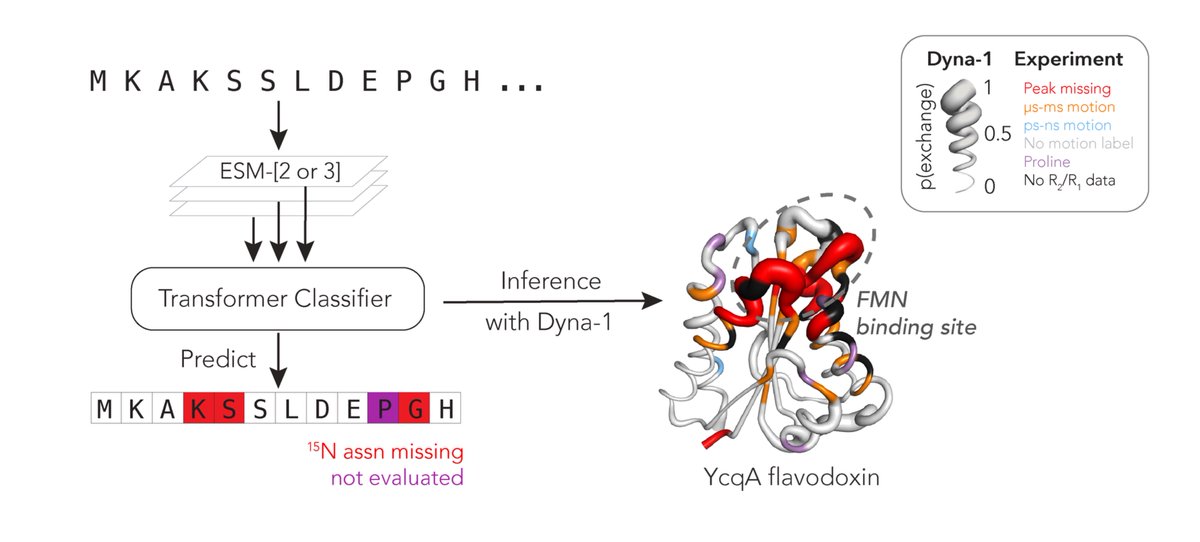
Protein function often depends on protein dynamics. To design proteins that function like natural ones, how do we predict their dynamics? Hannah Wayment-Steele and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1! 🧵