
Vadim Demichev
@demichevlab
Group Leader at @ChariteBerlin. Proteomics technologies and applications, DIA-NN author. aptila.bio
ID: 1058822152567304194
03-11-2018 20:44:07
1,1K Tweet
3,3K Takipçi
2,2K Takip Edilen



Excited to share our preprint on multiplexed DVP! mxDVP extends CODEX to DVP family, revealing spatial heterogeneity in human pancreatic islets with 12 endocrine sub-types including rare polyhormonal cells. Fantastic team effort Emma Lundberg #LundbergLab biorxiv.org/content/10.110…






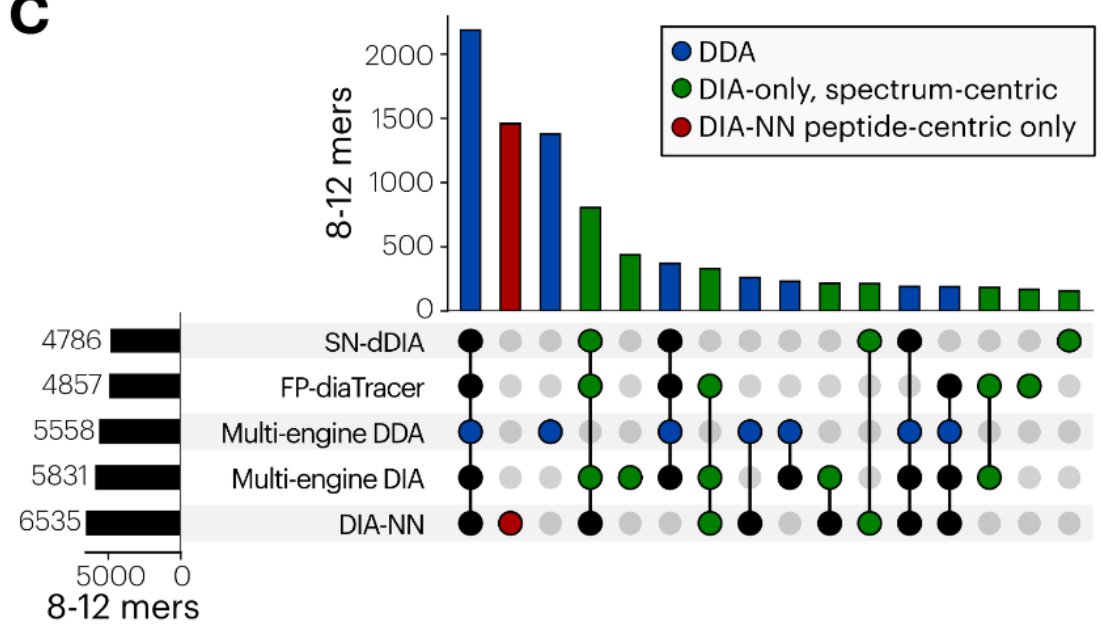
Was great to contribute to this fantastic work led by Patrick Willems. With the new proteoform confidence module in DIA-NN, we enabled non-specific digest searches. Here's how it now works for immunopeptidomics!



Join our #ASMS Breakfast Seminar on Monday, June 2, where Ashok Dongre, Senior Director and Head of #Proteomics at Bristol Myers Squibb, will share his latest research on the next generation of #Evosep chromatography workflows. Register now at evosep.com/asms2025 #teammassspec






Mathias Wilhelm, Martin Frejno from MSAID as well as Marie Locard-Paulet, Magnus Palmblad and myself are organising a workshop on FDR control & trustworthiness of proteomics, hosted by Lorentz Center in Leiden, February 2-6, 2026. Stay tuned :) lorentzcenter.nl/trustworthines…

I’m thrilled to share that the main work of my PhD has been published Cancer Cell! With Matthias Mann Lab & Ernst Lengyel, we analyzed the progression from ovarian serous borderline tumors to invasive cancer using spatial proteo-transcriptomics and identified new therapeutic targets.




