Dirk Benzinger
@dbenzingerd
Postdoc at @TheCrick in the Briscoe Lab. Previously PhD student with @KhammashLab. Dynamic signaling, gene regulation, synthetic biology, and optogenetics.
ID: 730369025755234304
11-05-2016 12:08:40
39 Tweet
159 Takipçi
397 Takip Edilen


Our work on synthetic signal-processing devices is now out authors.elsevier.com/a/1elT88YyDfj6…. We build optogenetic circuits that perform dynamic signal decoding and differential gene expression. Credit to Dirk Benzinger and S. Ovinnikov who did the work & European Research Council (ERC) ETH Zurich D-BSSE, ETH Zurich.


Very happy to see our paper on "synthetic circuits for dynamic signal processing/decoding" out. Thanks a lot to Serguei Ovinnikov who did a lot of great work on this during his masters thesis and of course to Mustafa Khammash for his support and supervision.

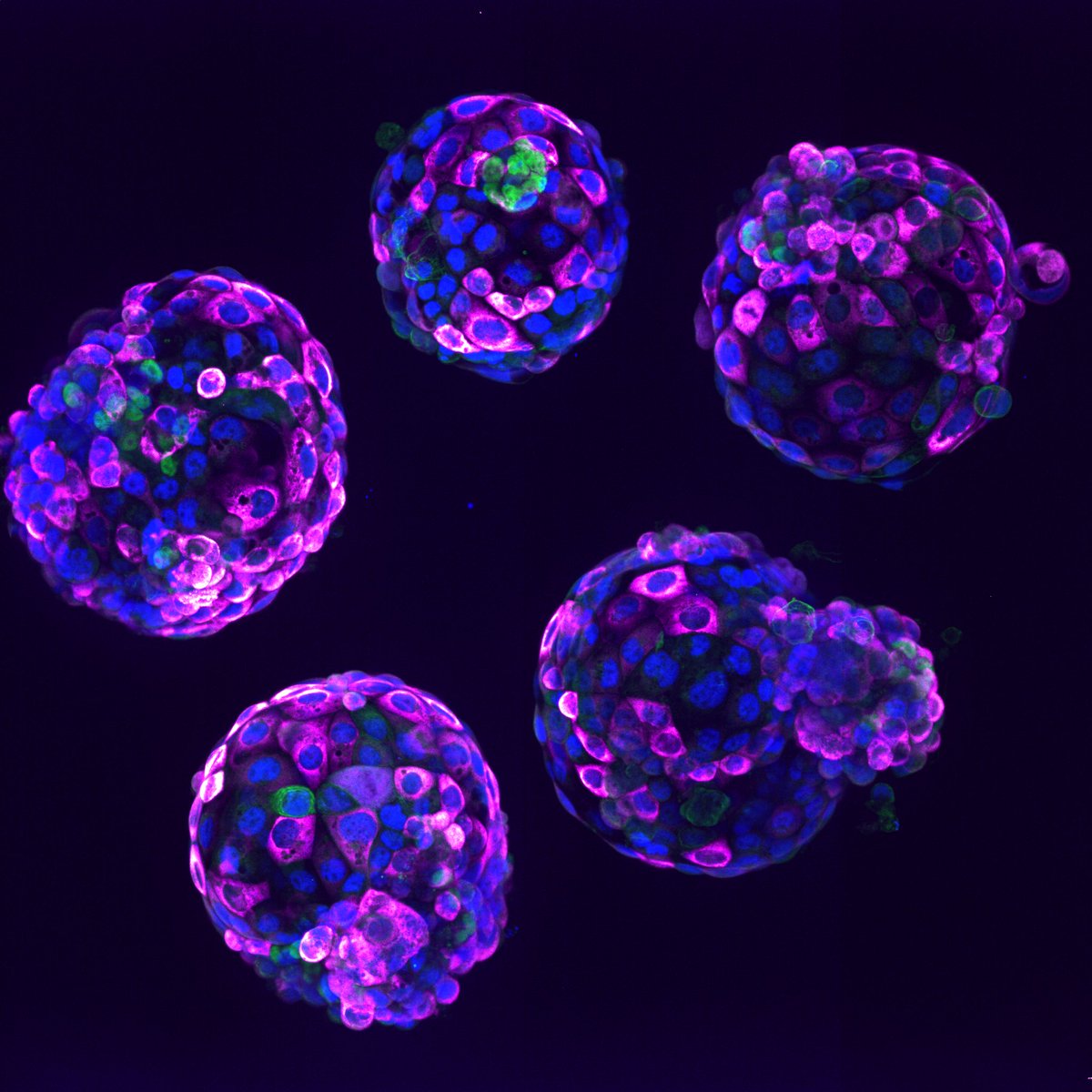
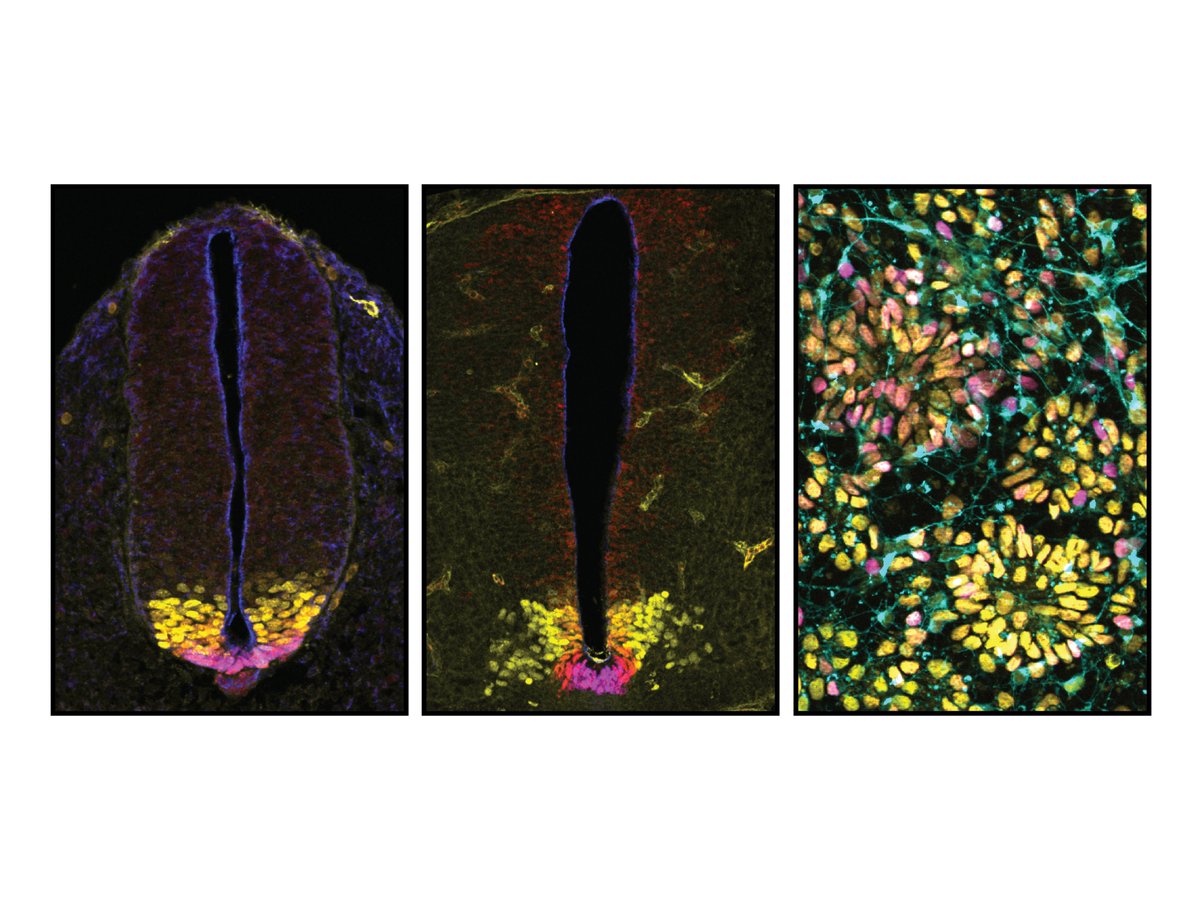
First project in James Briscoe lab. We uncover two cis regulatory strategies used in neural progenitor cell fate choice. It starts with the question: when you have one signal directing several alternative fates, what is the chromatin regulation strategy? tinyurl.com/ydbm76he

Come and work with us The Francis Crick Institute James Briscoe lab! Fixed-term Laboratory Research Officer (BSc or equivalent) Fun project understanding how cis regulatory elements control cell fate decisions in spinal cord development Closing 1st Aug! Please RT crick.wd3.myworkdayjobs.com/External/job/L…




Optogenetic feedback control of the unfolded protein response (UPR) in yeast can significantly increase the yield of a difficult-to-fold protein. #synbio #optogenetics authors.elsevier.com/a/1gl%7E4_Oxxo… Credit to Moritz@CTSB and Dirk Benzinger & thanks to ETH Zurich D-BSSE, ETH Zurich European Research Council (ERC)



Allison Saul explains how "Diamond IFFLs are a Signaling Decoder's Best Friend" in the latest UDS Writing Club post! Summary of a dazzling paper by Dirk Benzinger & Serguei Ovinnikov in the Mustafa Khammash 💎 kwrogers-lab.org/uds-writing-cl…





I am excited to share that I will be starting my lab at UCL as part of the LMCB UCL Laboratory for Molecular Cell Biology in 2024. We will be studying how cis-regulatory elements controls cell fate decisions during development. We are recruiting at all levels. Learn more and reach out!

🚨Meeting alert! Do you study cell lineages? In development, cancer, growth, regeneration? Join our The Royal Society meeting “Cell lineages across scales, space & time": barcoding, somatic tracing, imaging, modelling. Submit abstract, engage with leading speakers & win prizes!


🚨 The Delas lab UCL Laboratory for Molecular Cell Biology is officially recruiting 🥳 We will study how cis-regulatory elements control developmental cell fate choice, focusing on gene silencing. Funded by Wellcome. Postdoc advert closes on 12 March! ucl.ac.uk/work-at-ucl/se…