
Neil Thomas
@countablyfinite
Building AI for biological design @evoscaleai 🧬 Formerly: @Theteamatx 🌘 PhD @UCBerkeley 🙈
ID: 4105240514
https://thomas-a-neil.github.io/ 02-11-2015 20:04:05
400 Tweet
793 Takipçi
1,1K Takip Edilen

"Engineering of highly active and diverse nuclease enzymes by combining machine learning and ultra-high-throughput screening" from Neil Thomas Lucy Colwell and team looks amazing. They compare directed evolution, hit recombination, and ML-guided design of NucB.


Machine learning guided directed evolution beats directed evolution and hit recombination head-to-head for enzyme engineering. Neil Thomas Lucy Colwell





We're thrilled to present ESM3 in Science Magazine. ESM3 is a generative language model that reasons over the three fundamental properties of proteins: sequence, structure, and function. Today we're making ESM3 available free to researchers worldwide via the public beta of an API


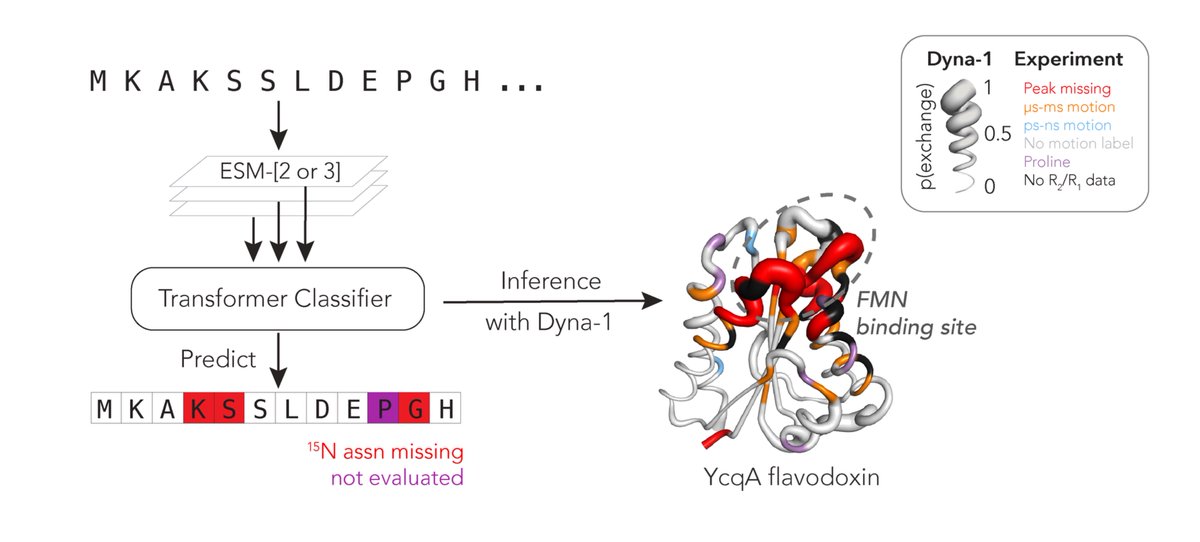
Protein function often depends on protein dynamics. To design proteins that function like natural ones, how do we predict their dynamics? Hannah Wayment-Steele and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1! 🧵

Gene synthesis is often the most expensive part of protein engineering with generative models. Happy to have played a small part in this work, where Chase developed a method for precision library construction at scale, with per-gene costs as low as $1.50. Romero lab


Thrilled to see my digital art on the cover of Trends in Genetics! The two binary strings represent reverse-complementary DNA sequences (00=A, 01=C, 10=G, 11=T) and the connecting rectangles represent “embeddings” learned by DNA language models. Our article: doi.org/10.1016/j.tig.…



